E5 POC and POR Species ID PCR and RFLPs
This post details protocls and results from identifying species of Porites and Pocillopora samples for the E5 RoL project.
Danielle Becker
Ariana Huffmyer
Overview
Previously, our lab has conducted Sanger sequencing of genetic markers to identify species within Porites and Pocillopora samples from the E5 RoL project using genetic markers.
Pocillopora species are identified using Sanger sequencing of the mtORF marker and a gel based RFLP of the POC Histone marker. Porites species are identified using Sanger sequencing of the H2 marker.
Protocols were developed by Zoe and Hollie based on previous work from Johnston et al. 2018 and Tisthammer et al. 2020.
Zoe and Hollie have previously processed samples using these protocols:
- Zoe’s PCR protocol is here and Species ID protocol is here.
- Zoe’s previous notebook posts using this protocol are here and here
- Hollie’s previous notebook posts using this protocol are here and protocol here.
After the processing detailed in Zoe’s notebooks, we need to run an RFLP assay on the Pocillopora samples to distinguish between P. meandrina and P. grandis in the samples that were distinguished from P. verrucosa through Sanger sequencing of the mtORF marker.
These samples are:
| Sample | Marker |
|---|---|
| POC-200 | POC Histone |
| POC-205 | POC Histone |
| POC-207 | POC Histone |
| POC-215 | POC Histone |
| POC-217 | POC Histone |
| POC-238 | POC Histone |
| POC-239 | POC Histone |
| POC-248 | POC Histone |
| POC-254 | POC Histone |
| POC-366 | POC Histone |
| POC-386 | POC Histone |
| POC-395 | POC Histone |
| POC-41 | POC Histone |
| POC-45 | POC Histone |
| POC-55 | POC Histone |
We also need to re run two samples of Porites that failed sequencing and/or amplification to distinguish between P. lutea/lobata and P. evermanni.
These samples are:
| Sample | Marker |
|---|---|
| POR-362 | Reamplify and resequence for H2 |
| POR-83 | Reamplify and resequence for H2 |
We will conduct PCR and RFLP for the Pocillopora samples (POC Histone) and will conduct PCR and Sanger sequencing for the Porites samples (H2).
Protocol
We are following Zoe’s Species ID PCR protocol here.
We will note below where we deviate from the standard protocol.
Equipment and supplies
- Sample Preservation and Lysis Zymo Research DNA/RNA Shield R1100-250
- DNA Extraction Zymo Research Quick-DNA™ Miniprep Plus Kit D4069
- Master Mix EmeraldAmp GT PCR Master Mix
- EmeraldAmp GT PCR Master Mix is a loading-dye-added version of EmeraldAmp MAX PCR Master Mix that is optimized for great performance and convenience in both standard and high-throughput PCR applications.
- Loading Dye NEB 6X Purple Loading Dye NEB Cat # B7024S
- Gel Stain Biotium GelGreen Nucleic Acid Gel Stain, 10,000X in Water Fisher Cat NC9728313
- DNA Ladder 1kb Gel ladder
- Gel ladder GeneRuler 100 bp DNA Ladder (Thermo FIsher Catalog SM0241)
- Restriction Enzyme XhoI R0146Sk
- Restriction Enzyme Buffer rCutSmart B6004S
- Forward primer (PocHistoneF: 5′-ATTCAGTCTCACTCACTCACTCAC- 3′) Johnston et al. 2018
- Reverse primer (PocHistoneR: 5′-TATCTTCGAACAGACCCACCAAAT-3′) Johnston et al. 2018
Note that we successfully ran this protocol on 20240329 because previous days protocols used the wrong primers. We were able to do some troubleshooting even so to improve this protocol. Scroll to 20240329 to see final results.
20240325
Pocillopora
Note that we ended up re doing this run on 20240326 because we added a positive control using a sample that was successfully amplified previously. The post for today details what we did, but the protocol did change slightly on subsequent days.
Danielle prepared samples and pulled the samples we will need to use from the freezer.
Sample metadata
| Sample ID | Tube No | Extracted Date |
|---|---|---|
| POC 200 | 273 | 20211007 |
| POC 205 | 289 | 20210924 |
| POC 207 | 309 | 20211105 |
| POC 215 | 335 | 20211115 |
| POC 217 | 81 | 20211015 |
| POC 238 | 321 | 20211018 |
| POC 239 | 317 | 20211104 |
| POC 248 | 325 | 20211007 |
| POC 254 | 311 | 20211019 |
| POC 366 | 363 | 20210916 |
| POC 386 | 465 | 20211005 |
| POC 395 | 447 | 20211122 |
| POC 41 | 407 | 20211015 |
| POC 45 | 383 | 20211001 |
| POC 55 | 391 | 20211118 |
| POR 362 | 707 | 20211020 |
| POR 83 | 469 | 20220208 |
15 POC samples
2 POR samples
1 negative control
Today we are just doing the POC samples, so there are 16 total to run.
POC Master Mix
We are planning the master mix for the POC samples for 16 samples ± 5-10% for error. We are planning for 18 total reactions to have enough master mix volume.
| Component | Concentration | 1 rxn (uL) | 18 rxns (uL) |
|---|---|---|---|
| EmeraldAmp 2X GT PCR Master Mix | 12.55 | 225.9 | |
| POC Histone F primer | 10 uM | 0.32 | 5.76 |
| POC Histone R primer | 10 uM | 0.32 | 5.76 |
| Ultra Pure H20 | 10.8 | 194.4 | |
| DNA | 1 | ||
| Total volume | 25uL |
Our primers are:
Poc Histone F Poc Histone R
POC PCR amplification
Master mix was made (without template DNA) by Danielle by first adding water, then primers, then the master mix.
Labeled PCR tubes with tube number in ascending order, with negative control as the last tube. Used PCR strip tubes.
Added 24uL of master mix made above to each tube. Added 1uL Ultrapure H20 as the negative control. Added 1 uL template DNA to each sample tube from each DNA extraction tube.
Samples added to the thermocycler at 11:30 for PCR, completed at 13:00.
The PCR protocol was as follows:
- [94°C 60 secondes] 1 cycle
- [94°C 30 sec,53°C 30 sec, 72°C 60 sec] 30 cycles
- [72°C 5 minutes] 1 cycle
- [4°C infinity]
- stored at 4°C until gel was run and we proceeded with restriction enzyme incubations
This PCR program is already programmed in the PPP lab thermocyclers.
POC Gel to QC PCR product
We next ran a gel to QC the PCR product. We prepared a 1% agarose gel in a medium box (20 wells) with 1XTAE buffer and 1uL of Gel Green added. Gel was run at 80V for 30 minutes from 14:56-15:26. 4uL of sample were added to each well with 2 ladders on either side (1KB Purple ladder 100uM).

The gel shows a consistent band length at 669bp as expected and the negative control is clear.
POC Restriction enzyme RFLP incubation
We then prepared the restriction enzyme master mix. We first diluted 10X rCutSmart to 1X (1uL rCutSmart and 9 uL Ultrapure H20). We planned for 20 reactions (18 samples + 10% error).
| Component | Concentration | 1 rxn | 20 rxns |
|---|---|---|---|
| rCutSmart | 1X | 0.5uL | 10uL |
| Xho1 RE | 0.5uL | 10uL | |
| Total | 20uL |
We then added 1uL of master mix and 15uL of PCR product into new strip tubes for each sample.
Tubes were loaded into a thermocycler and run for 1 h at 37°C followed by 20 min at 65°C with hold at 10°C.
POC Restriction enzyme RFLP gel
After the incubation, samples were removed and loaded into a 2% gel and ran at 70V for 1 h from 16:39-17:39.
There were no samples that were cut in this RFLP, which could mean they were all P. meandrina. However, without a positive control, we cannot be sure that the restriction enzymes worked, which could generating false identification of P. meandrina. We decided that tomorrow we will include previously amplified and RFLP’d samples from Hollie’s previous RFLP.
20240326
Pocillopora
POC Histone PCR
Today we re did samples ran yesterday but with a positive control. We are using sample POC-198 that successfully cut and was therefore identified as P. grandis in Hollie’s previous RFLP.
All other preparations an dprotocols were the same as described above, but with increased master mix volume for the one additional sample.
We started the PCR at 10:15 and it was completed by 11:50.
POC Gel PCR QC
We ran a 2% gel to QC POC Histone PCR product. We ran the gel at 80 V for 1h with 4uL of each sample and two ladders (same ladder as described above). Gel ran from 13:02-14:02.

We noticed that we have a band in our negative control. However, we think that this is a loading dye artifact because the same band is not seen in our other samples. We will try to fix this in our next round of PCR. The positive control sample amplified as did our other samples as expected.
POC Restriction enzyme RFLP incubation
We conducted the restriction enzyme incubation as done yesterday.
| Component | Concentration | 1 rxn | 20 rxns |
|---|---|---|---|
| rCutSmart | 1X | 0.5uL | 10uL |
| Xho1 RE | 0.5uL | 10uL | |
| Total | 20uL |
We then added 1uL of master mix and 15uL of PCR product into new strip tubes for each sample.
Tubes were loaded into a thermocycler and run for 1 h at 37°C followed by 20 min at 65°C with hold at 10°C from 13:32-15:00.
We also added another negative control (NEG2) that was the restriction enzyme master mix with 15 uL of H20.
POC RFLP gel
We ran a 2% gel at 80V for 1 h to view RFLP results. Gel ran from 13:45-14:45. We imaged the gel at 14:07. 1uL loading dye was added to NEG2 before adding to the gel.

We again saw no cutting in any of our samples and no cutting in the positive control. This suggests that that either one of two things is happening:
- The POC-198 colony is actually not P. grandis and is therefore not cutting.
- The restriction enzymes are bad or the protocol is not correct.
We checked the protocol multiple times against Hollie’s posts, Zoe’s posts, and the Johnston et al. 2018 paper and could not find any differences in our approach and other approaches. It is more likely that there is an issue with the restriction enzymes.
Porites
POR H2 PCR
Today we also started the process to PCR and prepare POR samples for Sanger sequencing using the H2 marker (see resources above).
We are planning the master mix for the POR samples for 2 samples, 2 positive controls, and 1 negative ± 5-10% for error. We are planning for 6 total reactions to have enough master mix volume.
| Component | Concentration | 1 rxn (uL) | 6 rxns (uL) |
|---|---|---|---|
| EmeraldAmp 2X GT PCR Master Mix | 12.55 | 75.3 | |
| zH2AH4f | 10 uM | 0.32 | 1.92 |
| zH4Fr R primer | 10 uM | 0.32 | 1.92 |
| Ultra Pure H20 | 10.8 | 64.8 | |
| DNA | 1 | ||
| Total volume | 25uL |
Our primers are:
zH2AH4f 5′-GTGTACTTGGCTGCYGTRCT-3′ zH4Fr 5′-GACAACCGAGAATGTCCGGT-3′
Our positive controls are:
- POR 209: P. lobata/lutea from the E5 time series previously sequenced (POS1)
- POR 72: P. evermanni from the E5 time series previously sequenced (POS2)
Our samples are:
- POR 362 (tube 707)
- POR 83 (tube 469)
We made the master mix and added 1uL template DNA to each tube, with 1uL H20 added to the negative control.
Samples were added to the thermocycler at 10:54-12:41. PCR was run using the same protocol as described above.
The PCR protocol was as follows:
- [94°C 60 secondes] 1 cycle
- [94°C 30 sec,53°C 30 sec, 72°C 60 sec] 30 cycles
- [72°C 5 minutes] 1 cycle
- [4°C infinity]
- stored at 4°C until prepared for sequencing
POR H2 QC Gel
We ran a 2% gel as described above for the POR H2 PCR product.

- We found that the POR 707 sample did not amplify (POR-362). This sample also did not amplify on previous attempts to sequence this sample.
- Both positive controls amplified and negative control was clear.
- 469 amplified from POR-83. In previous attempts for E5 processing this sample did amplify but failed sequencing. We will have to see what the sequencing result is to know if it was successful.
POR-362 was taken from tube 707 from time point 3. Notes in the E5 notebook say that this sample was taken from a physiology fragment, rather than a molecular tube. We will do another round of PCR where we take a different tube from this same colony from a different time point.
It could also be that DNA concentrations were too high or there are inhibitors present. To investigate this, we will also run diluted samples in our next round of PCR.
POR H2 PCR Attempt #2 today
We ran other PCR today and made two changes:
- We diluted sample 707 by 1/10 and 1/100
- We selected a new sample for colony POR-362 and added a sample of the full concentration, 1/10, and 1/100 dilutions.
We chose tube 837 as our new sample for POR-362. This sample is from time point 4 rather than time point 3.
We have 8 samples for this PCR:
- 707 diluted 1/10
- 707 diluted 1/100
- 837 full concentration
- 837 diluted 1/10
- 837 diluted 1/100
- POS1 (P. lobata/lutea sample 283)
- POS2 (P. evermanni sample 493)
- Negative control
We made a master mix, loaded samples into strip tubes for PCR as described above, and ran protocol from 16:46-18:31. PCR product was stored at 4°C overnight until, checking on a gel tomorrow morning.
20240327
Pocillopora
POC PCR
We ran a new round of PCR and RFLP today using three new positive control samples that were identified as P. grandis from POC spawning work in Moorea.
- POC-150 (POS1)
- POC-156 (POS2)
- POC-159 (POS3)
- POC-198 (POS4)
We chose to include POC-198 because it should be P. grandis, and we want to resolve this discrepancy.
We prepared the PCR as described above for 22 reactions and ran the PCR from 10:15-11:48.
POC PCR QC Gel
We ran another 2% gel as described above.

We had amplification as expected from our samples and the POC-198 positive control (POS4). Our negatives are clear. Unexpectedly, there was not much amplification of the POS1-3 controls. The band also appears to be shorter. It is possible that this specific region is a shorter sequence in P. grandis and that is why the bands are shorter. But if POC-198 is also P. grandis, it should have the same banding pattern and it does not - it is the same as our other samples.
POC RFLP incubation
We then took the PCR product, added the restriction enzyme master mix as described above, and incubated with restriction enzymes. We have 6 uL of PCR product left over after using 4uL for QC and 15uL for incubations. Left over PCR product was kept at 4°C.
Incubations took place from 12:45-14:05.
After this incubation, we ran another gel.
POC RFLP Gel QC
We ran a 2% gel from 14:44-15:44 for 1h at 80V as described above.

There was no cutting in any sample, even the positive controls. We concluded that the restriction enzyme is the problem. Either the reagents are degraded (which is unlikely with an expiration of 2025), or something is wrong with the master mix.
We looked back at Hollie’s notebooks and in her post she uses the same master mix and protocol that we did. But after talking with Hollie, she believes that she likely did not perform the dilution from 10X to 1X of the CutSmart buffer. In the case that a higher concentration is required of this buffer, we decided to do another RFLP incubation with 10X CutSmart rather than 1X to test this.
POC RFLP incubation with remaining PCR product using higher CutSmart concentration
We then prepared the following restriction enzyme master mix:
- 12 uL of Xhol
- 12 uL of 10X rCutSmart buffer
1uL of this mix was added to each remaining PCR product (6uL) along with 9uL ultrapure water (to bring sample volume to 15 uL). This resulted in a diluted PCR product, but because our bands are so bright, dilution should not reduce the concentration too low that we will not be able to see the banding pattern.
These were added to the thermocycler for incubation protocol at 17:20-18:40.
We then prepared a 2% gel and ran the gel at 80V for 50 min.

We did not see cutting in this gel. This means that the restriction enzyme is the problem.
We will run a new PCR tomorrow at a larger volume and with new enzymes that are scheduled to arrive tomorrow. If the enzymes don’t work again, we will prepare PCR product for sequencing as the alternative option.
Porites
POR PCR gel QC
We ran a 2% gel on yesterday’s PCR product at 70V for 1h at 10:20-11:20 as described above.

There was no amplification of sample 707 again, even with dilution. This sample may just be degraded. There was, however, amplification of sample 837 at all dilutions!
We will move forward with preparing the full concentration of 837 and 469 for Sanger sequencing.
PCR product clean up
Followed the exact protocol from the Zymo DNA Clean and Concentrator Kit
Buffer Preparation:
- Before starting: Add 24 ml 100% ethanol (26 ml 95% ethanol) to the 6 ml DNA Wash Buffer concentrate.
- Add 96 ml 100% ethanol (104 ml 95% ethanol) to the 24 ml DNA Wash Buffer concentrate.
Sample Processing:
- All centrifugation steps should be performed between 10,000 - 16,000 x g.
- In a 1.5 ml microcentrifuge tube, add 2-7 volumes of DNA Binding Buffer to each volume of DNA sample (see table below). Mix briefly by vortexing.
| Application | DNA Binding Buffer : Sample | Example |
|---|---|---|
| Plasmid, genomic DNA (>2 kb) | 2 : 1 | 200 µl : 100 µl |
| PCR product, DNA fragment | 5 : 1 | 500 µl : 100 µl |
| ssDNA (e.g. cDNA, M13 phage | 7 : 1 | 700 µl : 100 µl |
- We had 21 µl in our post PCR product tubes (25 µl reaction - 4 µl for post-PCR gel).
- We added 21 µl from each sample into a 1.5ml tube and added five times the initial volume 105 µl (21 µl x 5) of the DNA binding buffer to the 1.5 ml microcentrifuge tube.
- Transfer mixture to a provided Zymo-Spin™ Column in a Collection Tube.
- Centrifuge for 30 seconds. Discard the flow-through.
- Add 200 µl DNA Wash Buffer to the column. Centrifuge at for 30 seconds. Repeat the wash step.
- Add ≥ 25 µl DNA Elution Buffer directly to the column matrix and incubate at room temperature for one minute. Transfer the column to a 1.5 ml microcentrifuge tube and centrifuge at for 30 seconds to elute the DNA. Ultra-pure DNA is now ready for use.
NanoDrop
To check the purity of the POR DNA samples before sequencing, we used a NanoDrop on 20240427:
- I followed the NanoDrop Protocol exactly using the Zymo DNA Elution Buffer as my blank and the NanoDrop2000c in Dr. Jenkins lab CBLS 260.
Values:
| Sample ID | NanoDrop Concentration | 260/280 | 260/230 |
|---|---|---|---|
| POR_469 | 21.7 ng/uL | 2.17 | 0.65 |
| POR_837 | 39.8 ng/uL | 1.87 | 1.33 |
Composite Figures:
Preparation for Sanger sequencing
Danielle prepared the samples for Sanger sequencing for Janet at RI-INBRE.
First, dilute the two primer stock for sequencing (both F and R).
10 uM primer stock diluted to 3.2 uM:
1.6 uL of 10 uM primer + 3.4 uL H2) = 5 uL
Samples were diluted 1:1 with ultra pure H20 with 2uL H20 and 2uL PCR product
| Sample ID | Well (GSC use only) | Template type | A. Template size (bases) | B. Template stock conc. (ng/ul) | C. PCR template: ng needed = ((A/100)1.25)2 | D. PCR template: Volume=(C/B)ul | E. Plasmid template: volume= (200ng/B)*2 | F. Volume PCR H20 needed (10-D or E) | G. Volume primer needed 1ul per rxn | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Actual PCR product nanodop concentration | ||||||||||||
| Colony name | ||||||||||||
| 1 | PCR | 1500 | 10.85 | 37.5 | 3.5 | 6.5 | 2 | 21.7 | POR-469 | F | ||
| 2 | PCR | 1500 | 10.85 | 37.5 | 3.5 | 6.5 | 2 | 21.7 | POR-469 | R | ||
| 3 | PCR | 1500 | 19.9 | 37.5 | 1.9 | 8.1 | 2 | 39.8 | POR-837 | F | ||
| 4 | PCR | 1500 | 19.9 | 37.5 | 1.9 | 8.1 | 2 | 39.8 | POR-837 | R |
Danielle labeled the tubes and we will take them to Janet for sequencing tomorrow.
20240328
Pocillopora
Today we did a new round of PCR and RFLP with new restriction enzyme and buffer that arrived today.
Danielle ran this protocol today and Ariana ran the final gel.
POC PCR
We prepared a PCR as described previously using the new restriction enzymes. We have 20 samples with the 4 positive controls and 1 negative control. Danielle prepared master mix for 33 uL volume so that we have extra material to work with. We will plan for 22 reactions worth of master mix.
PCR master mix included:
| Component | Concentration | 1 rxn (uL) | 22 rxns (uL) |
|---|---|---|---|
| EmeraldAmp 2X GT PCR Master Mix | 16.66 | 366.52 | |
| F primer | 10 uM | 0.43 | 9.46 |
| R primer | 10 uM | 0.43 | 9.46 |
| Ultra Pure H20 | 14.66 | 322.52 | |
| DNA | 1 | ||
| Total volume | 33uL |
Each reaction included 32 uL master mix, 1uL template DNA, with ultra pure water added for negative controls.
The PCR started at 11:27 and ran until 13:04.
POC PCR QC gel
We ran a 2% gel at 80V for 1 h as described above with 4 uL for each sample.

E5 samples amplified, but POS4 is no longer amplifying. This is because we ran out of DNA and only a small amount was left for this reaction. The other positive controls 1 and 2 are showing a band. However, this band is slightly longer than our E5 samples. It is possible that these samples have a different amplified length of this region because we expect them to be P. grandis. We will see if this is the case when we get a successful RFLP.
POC RFLP incubation
We prepared a master mix of 1X rCutSmart buffer and Xhol RE with 12 uL of each as done previously. We added 1 uL of this master mix and 15 uL of sample to new strip tubes for RFLP incubation as done above. We added an additional water negative as we have before.
These samples ran from 15:35-16:35.
POC RFLP gel
We prepared at 2% gel and ran it at 80V for 1 hr from 18:05-19:05.

We did not see any cutting in this RFLP. Hollie talked to NEB today (the manufacturer of Xhol and rCutSmart) and asked them about the concentration of the rCutSmart buffer. In Zoe and Hollie’s protocols/posts and in the Johnston et al. 2018 paper, the buffer addition is referred to as “0.5 µL 1X CutSmart/rCutSmart buffer per reaction”. The stock of buffer comes in 10X concentration, so we have been reading this as the need to dilute the buffer before addition. In Zoe’s protocol, there is a dilution step to dilture the 10X buffer to 1X, which is then added as 0.5 µL per reaction.
However, as described yesterday, Hollie did not do this dilution in her protocol. Yesterday we tried adding the 10X buffer to the master mix rather than diluting but did not see any effect. It could be that this was because the buffer and/or restriction enzyme was degraded.
Today, Hollie talked with NEB and they said that the 1X refers to the concentration within the reaction, NOT the concentration before the addition to the master mix. Therefore, we will do one more RFLP tomorrow at the NEB suggested concentration and with the new buffer that arrived today.
Their recommendation is 5 µL buffer in a 50 µL reaction, which is a 1/10th dilution factor (5/50=0.1). This does not match the Johnson et al. concentrations or the concentrations in our protocol and is much higher than our current addition (0.5/15 = 0.033) and this will be 10 times less if we are doing the dilution before addition to the master mix if this is not required. Tomorrow we will run one more RFLP at the NEB suggested concentration with the remaining PCR product from today.
We will try this out and clarify in future posts and protocols whether the concentration of buffers is within the reaction or stock before the reaction.
We will use one of the following reaction volume options.
Full sample volume:
15 µL sample + 1.9 µL rCutSmart buffer + 1.9 µL Xhol = 18.8 µL total volume
1.9/18.8 = 0.10
Reduced sample volume (to save some for potential Sanger sequencing if RFLP doesn’t work):
7.5 µL sample + 0.95 µL rCutSmart buffer + 0.95 µL Xhol = 9.4 µL total volume
0.95/7.5 = 0.10
We are using the reduce sample volume master mix since we previously had success running RFLP with half sample volumes due to high concentration of amplicons after the PCR.
Porites
Danielle and Jill delivered the Porites prepared product for sequencing at RI-INBRE!
20240329
Pocillopora
POC RFLP incubation with 10X rCutSmart
We generated a master mix of 10X rCutSmart buffer and Xhol RE using the following volumes:
7.5 µL sample + 0.95 µL rCutSmart buffer + 0.95 µL Xhol = 9.4 µL total volume
We prepared a mix for 21 samples with 21.85 uL of each component. Each reaction included 7.5 uL of PCR product and 1.9 uL of master mix.
Samples were added to the thermocycler at 10:00-11:30 for the incubation.
During this time, we realized that the wrong primers were used (mtORF) instead of POC Histone! This now explains why the restriction enzymes weren’t cutting. We will now repeat the PCR and restriction enzyme incubation with the new primers.
PCR with correct POC Histone primers
Danielle ran a new PCR using the 33uL volumes as described above with the correct primers.
1 uL of template DNA was added to each sample.
| Component | Concentration | 1 rxn (uL) | 22 rxns (uL) |
|---|---|---|---|
| EmeraldAmp 2X GT PCR Master Mix | 16.66 | 366.52 | |
| F primer | 10 uM | 0.43 | 9.46 |
| R primer | 10 uM | 0.43 | 9.46 |
| Ultra Pure H20 | 14.66 | 322.52 | |
| DNA | 1 | ||
| Total volume | 33uL |
The PCR ran from 12:48-14:12.
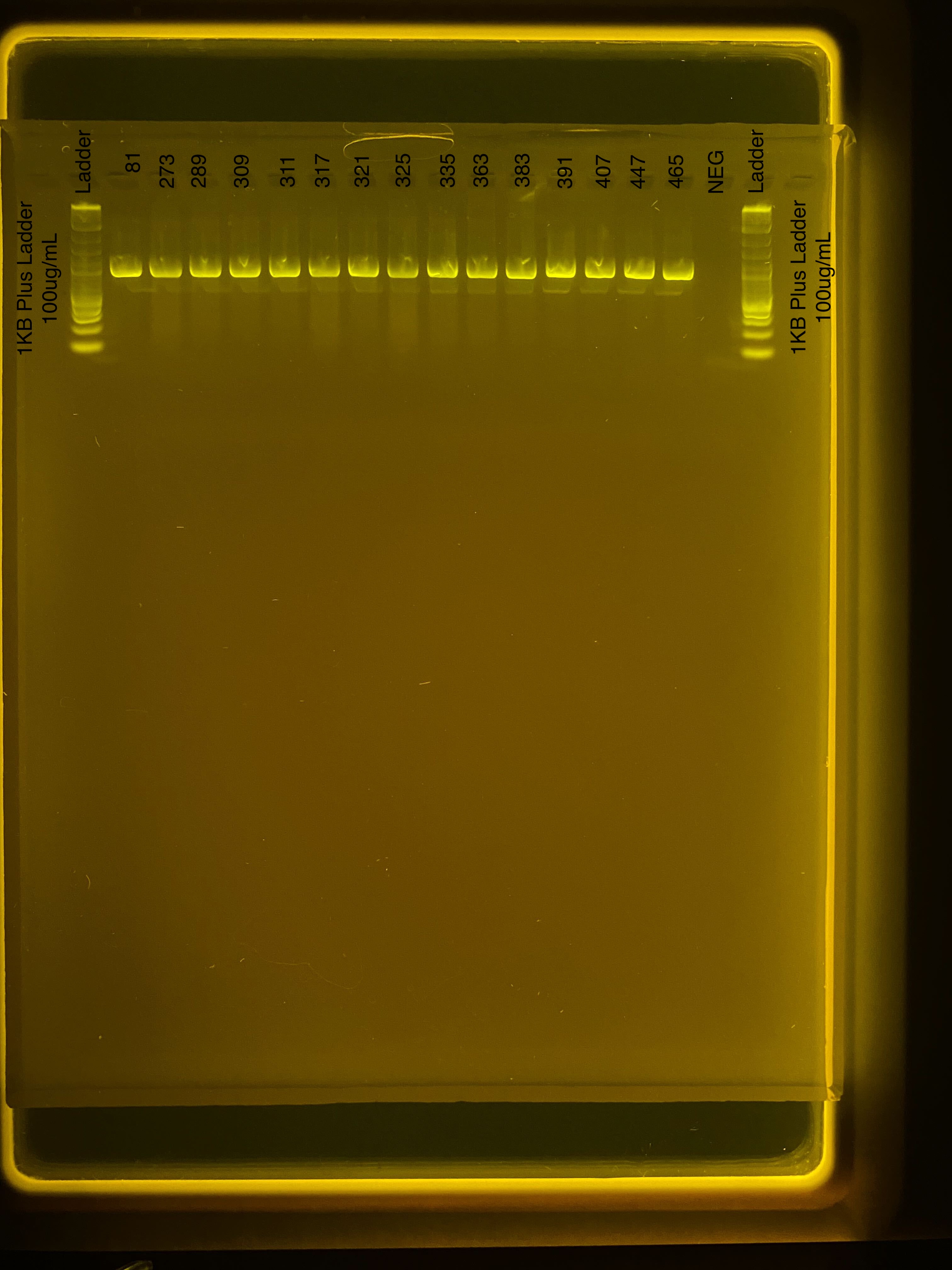
Gel of PCR product
We ran a gel at 80V for 1 hour with 3 uL of product. Gel ran from 14:50-15:50.

All samples amplified successfully, including all positives. Negatives are clear.
RFLP incubation
We prepared the master mix as we did yesterday at the higher concentration:
7.5 µL sample + 0.95 µL rCutSmart buffer + 0.95 µL Xhol = 9.4 µL total volume
Master mix was prepared with 21.85 uL of each component with 1.9 uL added to each reaction along with 7.5 uL of sample or water in the case of the negative.
Samples were added to the thermocycler at 15:33-16:53.
Gel of RFLP product
We prepared for a 2% gel and ran RFLP product at 80V for 1 hour with 3 uL sample in each well. The gel ran from 17:12-18:12.

It worked! All of our E5 samples are P. meandrina, shown by no cutting by the restriction enzyme. All four positive controls show successful cutting. Negative controls are clear.
RFLP and PCR products were discarded as they wont be used anymore.
Ariana and Danielle added sequencing data to the E5 GitHub and the Google Drive tracking sheet.
Results
All Pocillopora samples are P. meandrina and all Porites samples are P. evermanni.


