Oyster lifestage carryover project growth preliminary analysis
Post detailing preliminary analysis of oyster growth data in the USDA/WSG lifestage carry over project.
Overview
In this analysis, we looked at growth of oysters in the lifestage carry over project, which is detailed in Eric’s notebook here.
In this post I detail preliminary growth analyses. For this experiment, we have pools of lifestages in different containers, represented as replicates (n=1-4 per treatment). For adults, we had individual families tracked by tag number. We did not identify or track individual oysters and do not have replicates within individual tags. Therefore, we treat tag/container as replicates within treatment and will only have limited capacity to make inferences about family effects.
Setup
Set up workspace, set options, and load required packages.
knitr::opts_chunk$set(echo = TRUE, warning = FALSE, message = FALSE)
Load libraries
library(tidyverse)
library(ggplot2)
Read in data
growth<-read_csv("lifestage_carryover/data/size/gigas-lengths.csv")
growth<-growth%>%
rename(tag=`tag...1`)%>%
select(tag, `life-stage`, `init-treat`, label, month, length)%>%
rename(lifestage=`life-stage`, treatment=`init-treat`)
growth$month<-factor(growth$month, levels=c("Oct", "Nov", "Jan"))
Look at data for whole population together
Plot growth over months for each life stage and treatment
Plot as box plot
plot<-growth%>%
ggplot(aes(color=treatment, x=month, y=length))+
geom_boxplot()+
facet_grid(~lifestage)+
ylab("Mean Length (cm)")+
xlab("Month")+
scale_color_manual(values=c("darkgray", "darkred"), name="Treatment", labels=c("Control", "Treatment"))+
theme_classic()+
theme(
text=element_text(size=12, color="black")
);plot
ggsave(plot=plot, "lifestage_carryover/output/size/mean_lengths_box.png", width=7, height=4)

Plot as mean +- standard error for all.
plot2<-growth%>%
group_by(treatment, month, lifestage)%>%
summarise(mean=mean(length, na.rm=TRUE), se=sd(length, na.rm=TRUE)/sqrt(length(length)))%>%
ggplot(aes(color=treatment, x=month, y=mean))+
geom_point(aes(group=interaction(treatment, lifestage)), position=position_dodge(0.3), size=1)+
geom_errorbar(aes(ymin=mean-se, ymax=mean+se), width=0, position=position_dodge(0.3), linewidth=0.75)+
geom_line(aes(group=treatment), position=position_dodge(0.3), linewidth=1)+
facet_grid(~lifestage)+
ylab("Mean Length (cm)")+
xlab("Month")+
scale_color_manual(values=c("darkgray", "darkred"), name="Treatment", labels=c("Control", "Treatment"))+
theme_classic()+
theme(
text=element_text(size=12, color="black")
);plot2
ggsave(plot=plot2, "lifestage_carryover/output/size/mean_lengths.png", width=7, height=4)

Run model
model<-aov(length~lifestage*month*treatment, data=growth)
summary(model)
Significant lifestage and month effects. No significant treatment effects.
Df Sum Sq Mean Sq F value Pr(>F)
lifestage 3 383973 127991 1377.056 <2e-16 ***
month 2 9529 4765 51.264 <2e-16 ***
treatment 1 157 157 1.686 0.194
lifestage:month 3 15429 5143 55.335 <2e-16 ***
lifestage:treatment 3 510 170 1.830 0.140
month:treatment 2 15 8 0.081 0.922
lifestage:month:treatment 3 26 9 0.094 0.963
Residuals 1006 93503 93
Run plot and model by tag
Calculate mean size at each month for each replicate and calculate % growth (change/original).
replicates<-growth%>%
group_by(treatment, month, lifestage, tag)%>%
summarise(mean=mean(length, na.rm=TRUE))%>%
pivot_wider(names_from=month, values_from=mean)%>%
mutate(perc_growth_Nov=((Nov-Oct)/Oct)*100)%>%
mutate(perc_growth_Jan=((Jan-Oct)/Oct)*100)
Plot growth for adult animals sampled in November
plot3<-replicates%>%
select(treatment, lifestage, tag, perc_growth_Nov)%>%
drop_na()%>%
droplevels()%>%
ggplot(aes(color=treatment, x=treatment, y=perc_growth_Nov, shape=tag))+
geom_line(aes(group=tag), color="black")+
geom_point(aes(group=interaction(treatment, lifestage)), position=position_dodge(0.3), size=5)+
ylab("% growth")+
xlab("Treatment")+
scale_color_manual(values=c("darkgray", "darkred"), name="Treatment", labels=c("Control", "Treatment"))+
scale_shape_manual(name="Family", values=c(15, 16, 17, 18))+
ggtitle("Adults: Oct-Nov Growth")+
theme_classic()+
theme(
text=element_text(size=12, color="black")
);plot3
ggsave(plot=plot3, "lifestage_carryover/output/size/adults_Nov_percent_growth.png", width=4, height=4)

Test for effect of treatment.
model<-replicates%>%
select(treatment, lifestage, tag, perc_growth_Nov)%>%
drop_na()%>%
droplevels()%>%
aov(perc_growth_Nov~treatment, data=.)
summary(model)
Effect of treatment just barely not significant (P=0.088).
Df Sum Sq Mean Sq F value Pr(>F)
treatment 1 8.358 8.358 4.132 0.0883 .
Residuals 6 12.137 2.023
Plot growth for animals measured in January
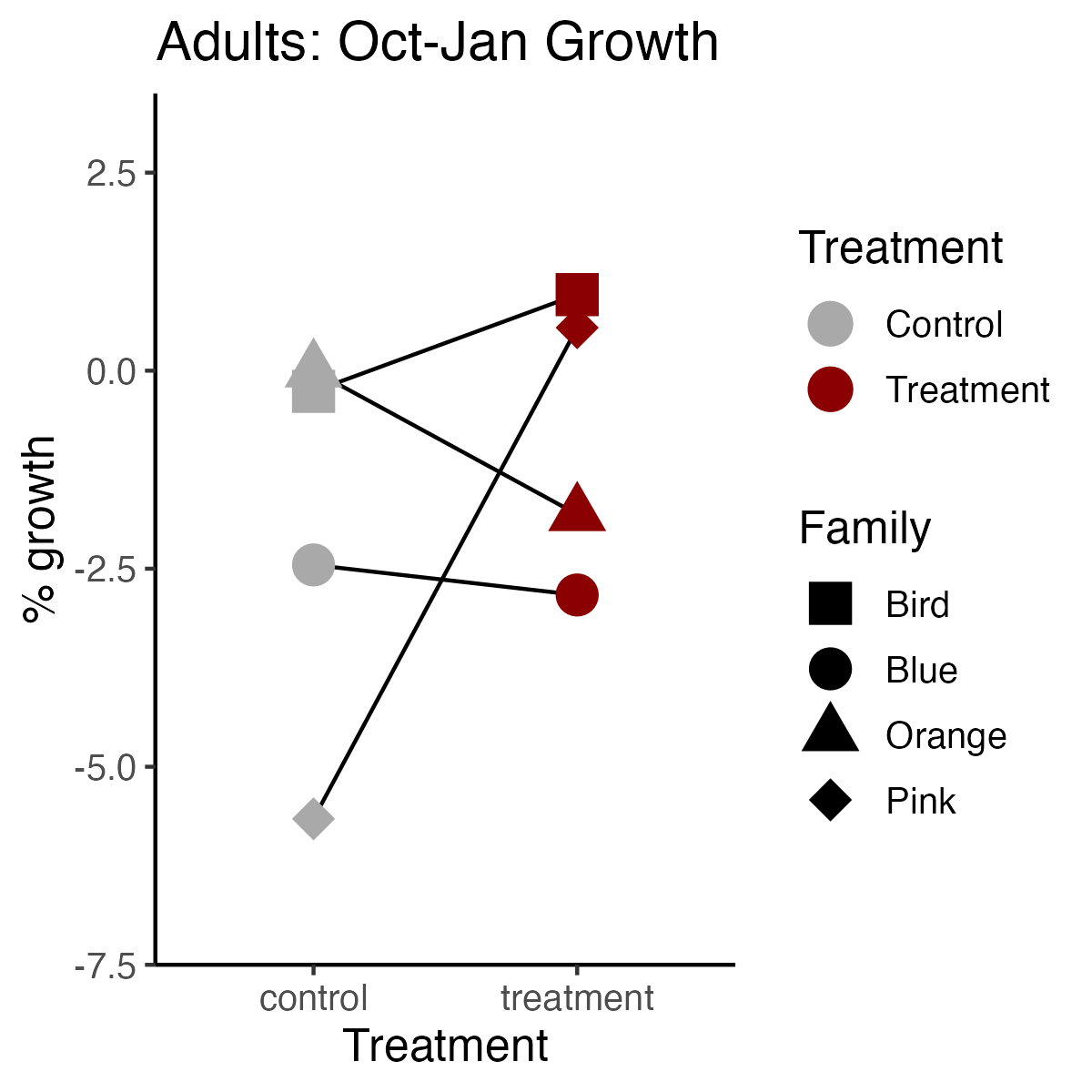
Adults
plot4<-replicates%>%
select(treatment, lifestage, tag, perc_growth_Jan)%>%
filter(lifestage=="adult")%>%
drop_na()%>%
droplevels()%>%
ggplot(aes(color=treatment, x=treatment, y=perc_growth_Jan, shape=tag))+
geom_line(aes(group=tag), color="black")+
geom_point(aes(group=interaction(treatment, lifestage)), position=position_dodge(0.3), size=5)+
ylab("% growth")+
xlab("Treatment")+
scale_color_manual(values=c("darkgray", "darkred"), name="Treatment", labels=c("Control", "Treatment"))+
ylim(-7, 3)+
scale_shape_manual(name="Family", values=c(15, 16, 17, 18))+
ggtitle("Adults: Oct-Jan Growth")+
theme_classic()+
theme(
text=element_text(size=12, color="black")
);plot4
ggsave(plot=plot4, "lifestage_carryover/output/size/adults_Jan_percent_growth.png", width=4, height=4)
Juveniles
plot5<-replicates%>%
select(treatment, lifestage, tag, perc_growth_Jan)%>%
filter(lifestage=="juvenile")%>%
drop_na()%>%
droplevels()%>%
ggplot(aes(color=treatment, x=treatment, y=perc_growth_Jan, shape=tag))+
geom_point(aes(group=interaction(treatment, lifestage)), position=position_dodge(0.3), size=5)+
ylab("% growth")+
xlab("Treatment")+
scale_color_manual(values=c("darkgray", "darkred"), name="Treatment", labels=c("Control", "Treatment"))+
scale_shape_manual(name="Replicate", values=c(15, 16, 17, 18))+
ggtitle("Juvenile: Oct-Jan Growth")+
theme_classic()+
ylim(0,20)+
theme(
text=element_text(size=12, color="black")
);plot5
ggsave(plot=plot5, "lifestage_carryover/output/size/juvenile_Jan_percent_growth.png", width=4, height=4)

Seed
plot6<-replicates%>%
select(treatment, lifestage, tag, perc_growth_Jan)%>%
filter(lifestage=="seed")%>%
drop_na()%>%
droplevels()%>%
ggplot(aes(color=treatment, x=treatment, y=perc_growth_Jan, shape=tag))+
#geom_line(aes(group=tag), color="black")+
geom_point(aes(group=interaction(treatment, lifestage)), position=position_dodge(0.3), size=5)+
ylab("% growth")+
xlab("Treatment")+
scale_color_manual(values=c("darkgray", "darkred"), name="Treatment", labels=c("Control", "Treatment"))+
scale_shape_manual(name="Replicate", values=c(15, 16, 17, 18))+
ggtitle("Seed: Oct-Jan Growth")+
theme_classic()+
ylim(0,250)+
theme(
text=element_text(size=12, color="black")
);plot6
ggsave(plot=plot6, "lifestage_carryover/output/size/seed_Jan_percent_growth.png", width=4, height=4)

Spat
plot7<-replicates%>%
select(treatment, lifestage, tag, perc_growth_Jan)%>%
filter(lifestage=="spat")%>%
drop_na()%>%
droplevels()%>%
ggplot(aes(color=treatment, x=treatment, y=perc_growth_Jan, shape=tag))+
#geom_line(aes(group=tag), color="black")+
geom_point(aes(group=interaction(treatment, lifestage)), position=position_dodge(0.3), size=5)+
ylab("% growth")+
xlab("Treatment")+
scale_color_manual(values=c("darkgray", "darkred"), name="Treatment", labels=c("Control", "Treatment"))+
scale_shape_manual(name="Replicate", values=c(15, 16, 17, 18))+
ggtitle("Spat: Oct-Jan Growth")+
theme_classic()+
ylim(0,150)+
theme(
text=element_text(size=12, color="black")
);plot7
ggsave(plot=plot7, "lifestage_carryover/output/size/spat_Jan_percent_growth.png", width=4, height=4)

Test for effect of treatment by lifestage
January all stages
model<-replicates%>%
select(treatment, lifestage, tag, perc_growth_Jan)%>%
drop_na()%>%
droplevels()%>%
aov(perc_growth_Jan~treatment*lifestage, data=.)
summary(model)
Effect of lifestage, no treatment effects.
Df Sum Sq Mean Sq F value Pr(>F)
treatment 1 1 1 0.015 0.904
lifestage 3 96056 32019 325.275 3.54e-13 ***
treatment:lifestage 3 363 121 1.231 0.335
Residuals 14 1378 98
Interpretations
There are a few key take aways here:
- Adults (1/2 of families) that were sampled in November were measured for growth in November. These families tended to be larger, so that is why lengths of adults are higher in November. All other adults and other lifestages were measured in January.
- The amount of growth is much higher on a percent basis in seed and spat than adults and juveniles. This demonstrates lifestage specific investment in growth and could be interesting to explore further.
- There are no strong treatment effects. There are some trends for family specific treatment effects in adults, but because we do not have replicates we cannot make any conclusions on adult family effects.


