Chlorophyll Content Protocol
This protocol details chlorophyll measurement in coral larval tissue samples. This protocol is modified from the E5 chlorophyll protocol.
Note that this protocol needs to be optimized prior to use for ASH larval samples
Quantifying Chl-a and Chl-c2 Concentration in Symbiodiniaceae from coral tissues
Original: 20200101
Last Revised: 20220413 ASH
Contents
- Materials
- 100% acetone
- flammable safe fridge 4°
- quartz 96 well plate
- Microcentrifuge (Mo’orea Gump Molecular Lab)
- 1ml pipette and tips
- 1.5 ml microfuge tubes
- Synergy HTX Multi-Mode Microplate Reader (PPP lab plate reader)
- Gen5 Software (PPP Lab computer)
Sample Preparation
- Thaw homogenate aliquot. These samples are previously separated using the fraction separation protocol described here.
- Centrifuge the aliquot of larval homogenate at 13,000 rpm for 3 minutes to separate the host and Symbiodiniaceae cells.
- Remove the supernatant and add to a new tube labeled “Holo-Supernatant”.
- Add 1 mL of 100% acetone to the pellet in the 1.5 mL microcentrifuge tube and vortex the tubes for 15 sec.
- Place the tubes in a fridge in the dark at 4°C for 24 hours.
- Vortex the tubes for 15 sec.
- Spin the tubes down at 13,000 rpm for 3 minutes in the microcentrifuge to pellet any debris.
- Pipette 200µl of sample to duplicate wells of 96-well quartz plate.
- Pipette 200µl of acetone blank to duplicate wells.
- Cover the plate with silicone pad every 5th sample or so to reduce evaporate as samples are added.
- Remove silicon pad.
- Follow steps below to measure the extract Absorbance on the Synergy HTX Multi-Mode Microplate Reader at 630, 663, and 750 nm in a 96-well quartz plate.
- Standardize for path length in 200µl of sample in 96-well quartz plate.
Measure the Absorbance
Follow the Synergy HTX Operating Manual and Gen5 Software Manual to install the software on your host computer and general operating instructions.
- Open the Gen5 software on your computer.
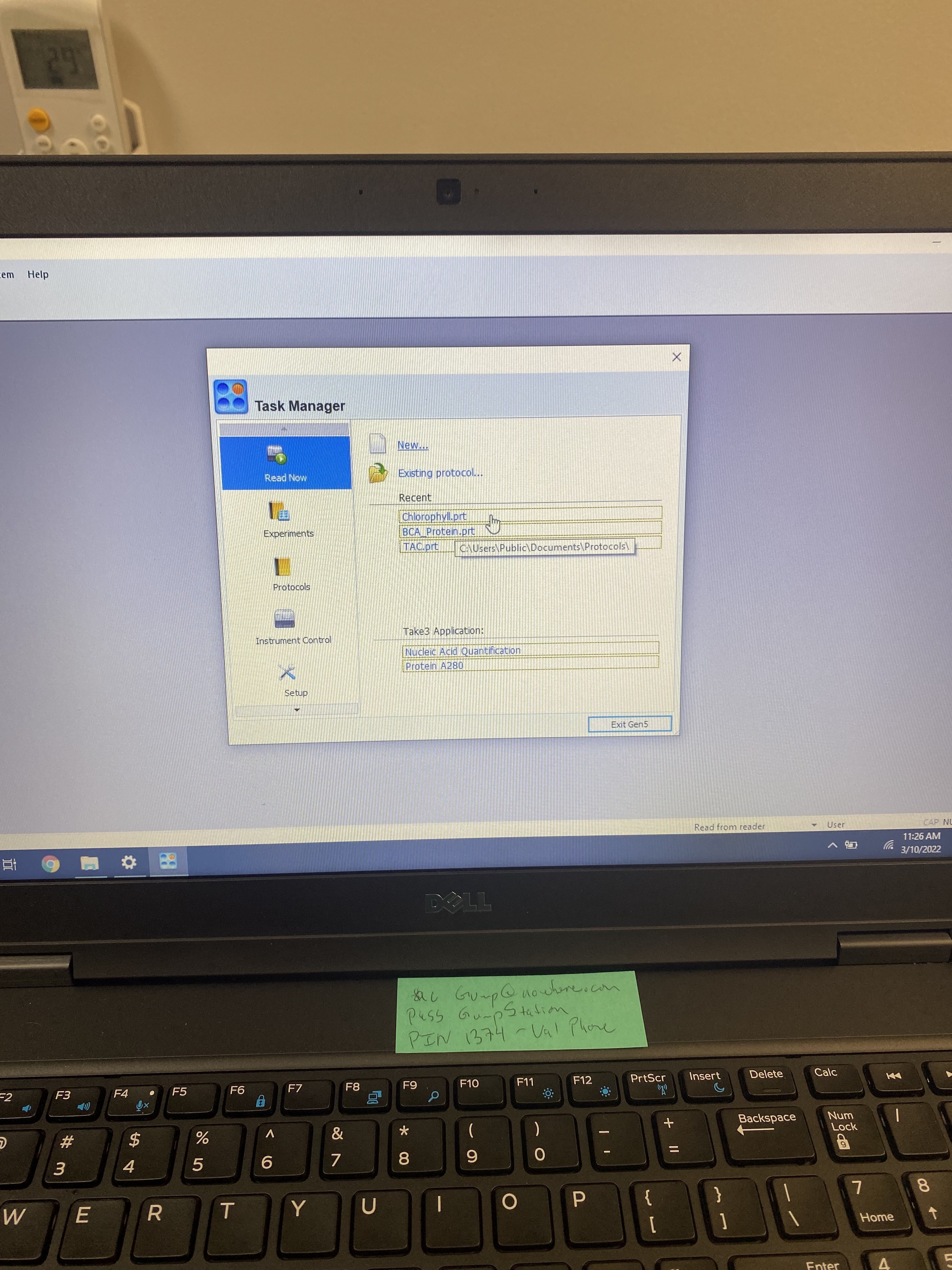
- Whenever you start Gen5, the Task Manager opens. Load the presaved chlorophyll protocol on the Gen5 software on the HP Putnam Lab computer.
- To create a new protocol, follow the ‘Getting Started’ section in the Gen5 Software Manual.
- The plate loader should automatically open. Load your plate following the distinctions in the plate loader and you will be all set.
- To measure Absorbance, select the ‘Chlorophyll Protocol’ option.
- Press the green start protocol button in the GEN5 software and allow the protocol to run through the 630, 663, and 750 nm.
- Export the raw GEN5 software data file and a CSV to the Desktop in a designated folder. Also, immediately upload to a google drive folder to have for future use.
- Standardize for path length in 200µl of sample in 96-well quartz plate.
Calculating Chlorophyll Concentration
Chlorophyll a and c2 concentrations are calculated from the equations in Jeffrey and Humphrey 1975 after substracting A750nm from all measurements.
Need to correct for differences in path length of the volume in the 96 well plate compared to the 1cm path length of a cuvette. Warren 2007


