Mcapitata Development 16S Analysis Part 1
This post details MultiQC for the 16S analysis pipeline adapted from the pipeline developed by Emma Strand. In this post there is information for moving/managing ITS2 sequences, but this post details preliminary analysis of 16S. ITS2 files will be analyzed in a separate post.
16S MultiQC
1. Log onto Andromeda and go to working directory
Log into Andromeda
ssh -l ashuffmyer ssh3.hac.uri.edu
Enter password, then navigate to working directory.
cd /data/putnamlab/ashuffmyer
Copy files from shared drive to working directory. Each sample will have an R1 and R2 fastq.gz files.
cp -r /data/putnamlab/shared/AH_MCAP_ITS2 /data/putnamlab/ashuffmyer
cp -r /data/putnamlab/shared/AH_MCAP_16S /data/putnamlab/ashuffmyer
cd AH_MCAP_16S
2. Create a conda environment
Download miniconda and then create a conda environment.
Load modules that are required.
module load Python
module load Anaconda3
module load Miniconda3
Verify that you are working with Miniconda.
which python, this should return Miniconda version.
Create a conda environment.
conda create -n AH_MCAP_16S and enter y for proceed.
Activate the environment.
conda activate AH_MCAP_16S
When done working, use conda deactivate AH_MCAP_16S
Note that you have to activate the environment each time you work in this environment.
If needed, initiate conda for your shell, in this case I am using bash.
conda init bash
After activating the environment, the line should now look like this:
(AH_MCAP_16S) [ashuffmyer@ssh3 AH_MCAP_16S]$
3. Check that programs are available
Check for available modules that will be needed. If needed, contact URI to ask for modules to be added.
E. Strand pipeline uses the following:
module load Miniconda3/4.9.2
module load FastQC/0.11.9-Java-11
module load MultiQC/1.9-intel-2020a-Python-3.8.2
module load cutadapt/2.10-GCCcore-9.3.0-Python-3.8.2
module load QIIME2/2021.4
4. Run FASTQC for quality control of raw read files
Information on interpretting fastQC results here.
Create a report of all FastQC using MultiQC.
Create a new directory for the fastqc results.
mkdir fastqc_results
cd fastqc_results
Move raw data files into a directory for ease of navigating the directory.
cd ..
mkdir raw_data
Move all .gz files into the new raw_data directory.
mv /data/putnamlab/ashuffmyer/AH_MCAP_16S/*.gz /data/putnamlab/ashuffmyer/AH_MCAP_16S/raw_data
Make a folder for scripts.
mkdir scripts
Write a script to run the fastqc.
cd scripts
nano fastqc.sh
This script will run on the putnamlab node with updates sent via email. Note the directories - this script lives in scripts and directories are reflected from this location.
#!/bin/bash
#SBATCH -t 24:00:00
#SBATCH --nodes=1 --ntasks-per-node=1
#SBATCH --export=NONE
#SBATCH --mem=100GB
#SBATCH --mail-type=BEGIN,END,FAIL #email you when job starts, stops and/or fails
#SBATCH --mail-user=ashuffmyer@uri.edu #your email to send notifications
#SBATCH --account=putnamlab
#SBATCH --error="fastqc_script_error" #if your job fails, the error report will be put in this file
#SBATCH --output="fastqc_output_script" #once your job is completed, any final job report comments will be put in this file
source /usr/share/Modules/init/sh # load the module function
module load FastQC/0.11.9-Java-11
module load MultiQC/1.9-intel-2020a-Python-3.8.2
for file in ../raw_data/*fastq.gz
do
fastqc $file --outdir ../fastqc_results
done
multiqc --interactive ../fastqc_results
mv multiqc_report.html ../fastqc_results/16S_raw_qc_multiqc_report_AH.html #renames file
Run the script.
sbatch fastqc.sh
Job will submit. Check the status of a job by typing squeue -u ashuffmyer. This will show the status of all jobs by you as a user. For 78 files, this took about 8-10 minutes.
If you need to troubleshoot with analyzing single files, use interactive and exit to quit interactive mode.
To view script errors use nano script_error.
Check that all files were processed.
cd ../raw_data
ls -1 | wc -l
This is the number of raw files that we have. There are 78 files for this project (39 samples).
cd ../fastqc_results
ls -1 | wc -l
This should be 78 x 2 = 156 (zip and html file for each raw data file). This was correct.
5. MultiQC report: Visualizing FastQC
Copy the report to home desktop. Open a terminal session outside of Andromeda and use login credentials when prompted.
scp ashuffmyer@bluewaves.uri.edu:/data/putnamlab/ashuffmyer/AH_MCAP_16S/fastqc_results/16S_raw_qc_multiqc_report_AH.html ~/MyProjects/EarlyLifeHistory_Energetics/Mcap2020/Data/16S
Initial MultiQC report ran on 20211227 shows several areas that we need to address. MultiQC report including:
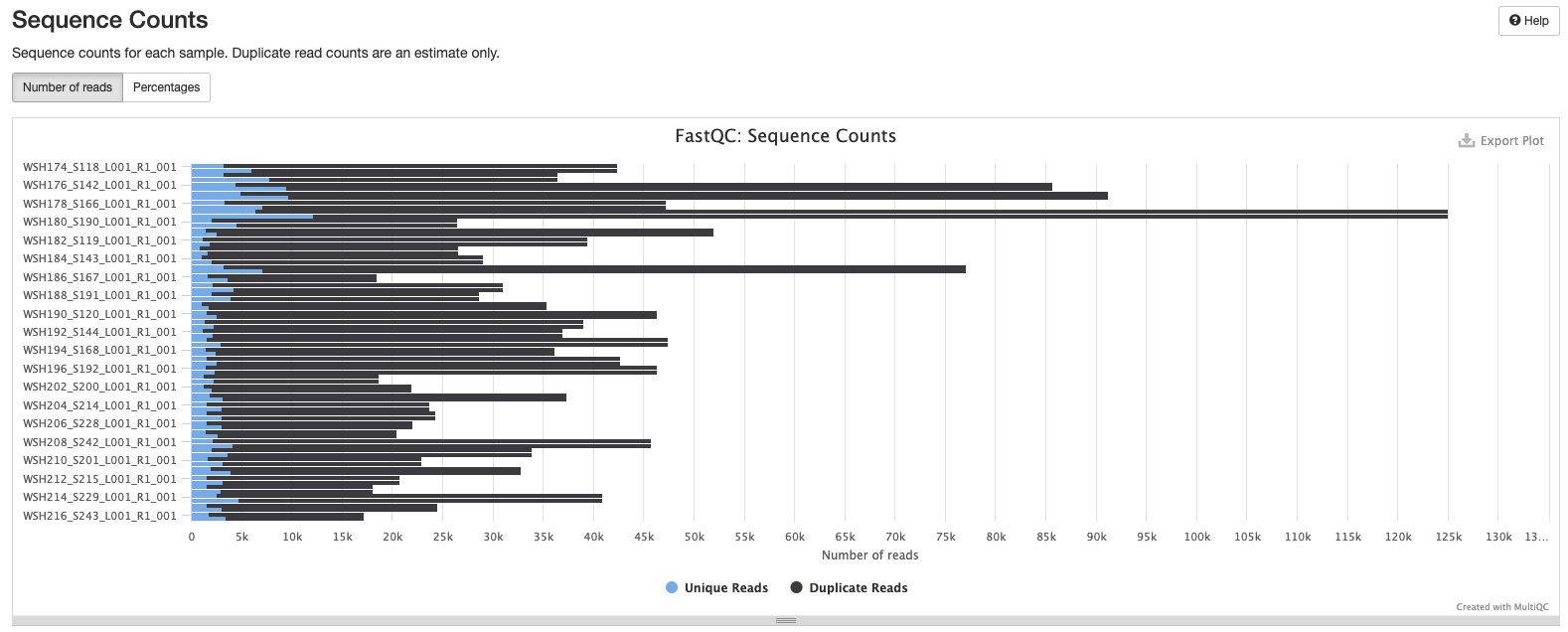
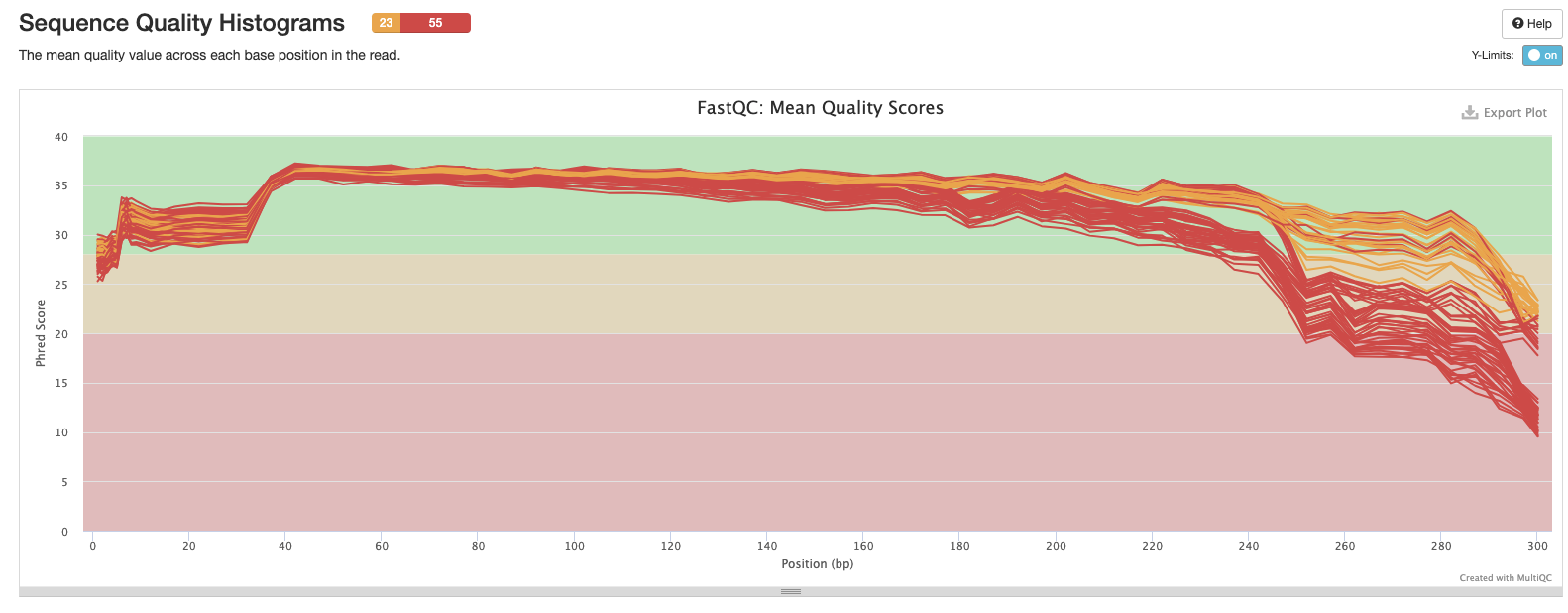
Interpretation: Reduction in quality at the end of the sequence.
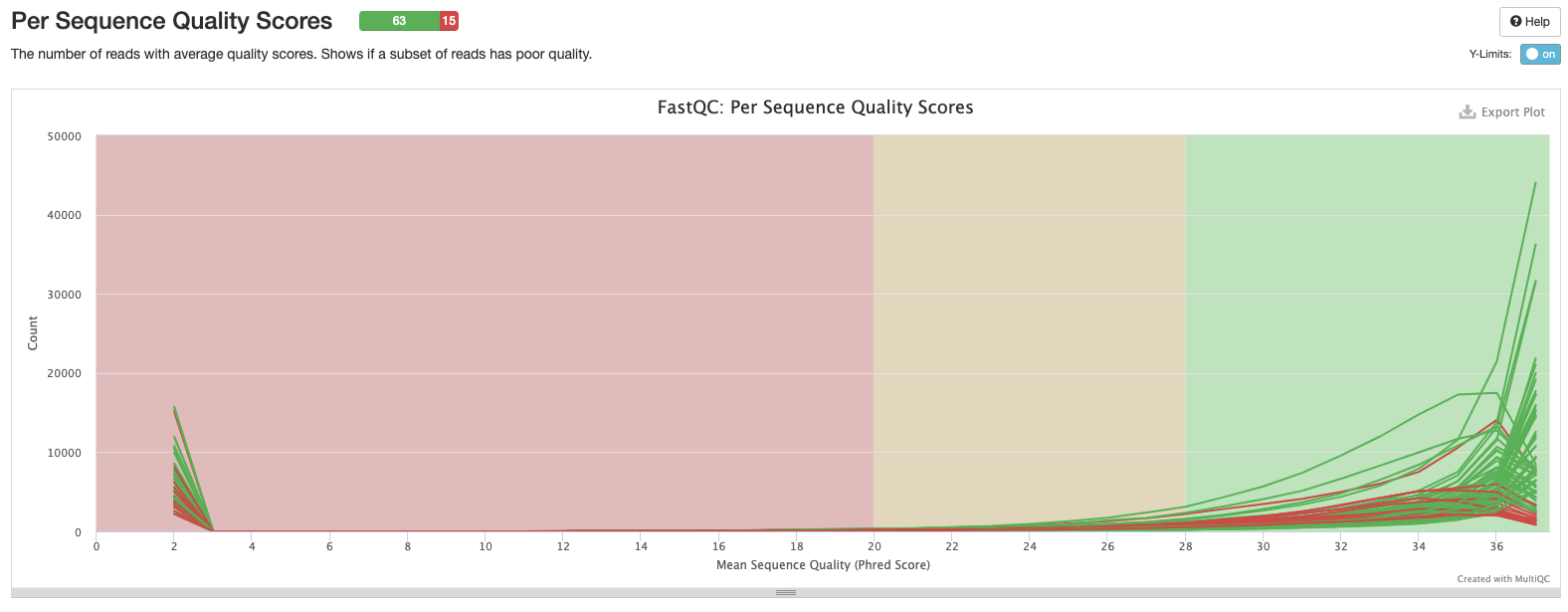
Interpretation: 15 samples have low quality scores.
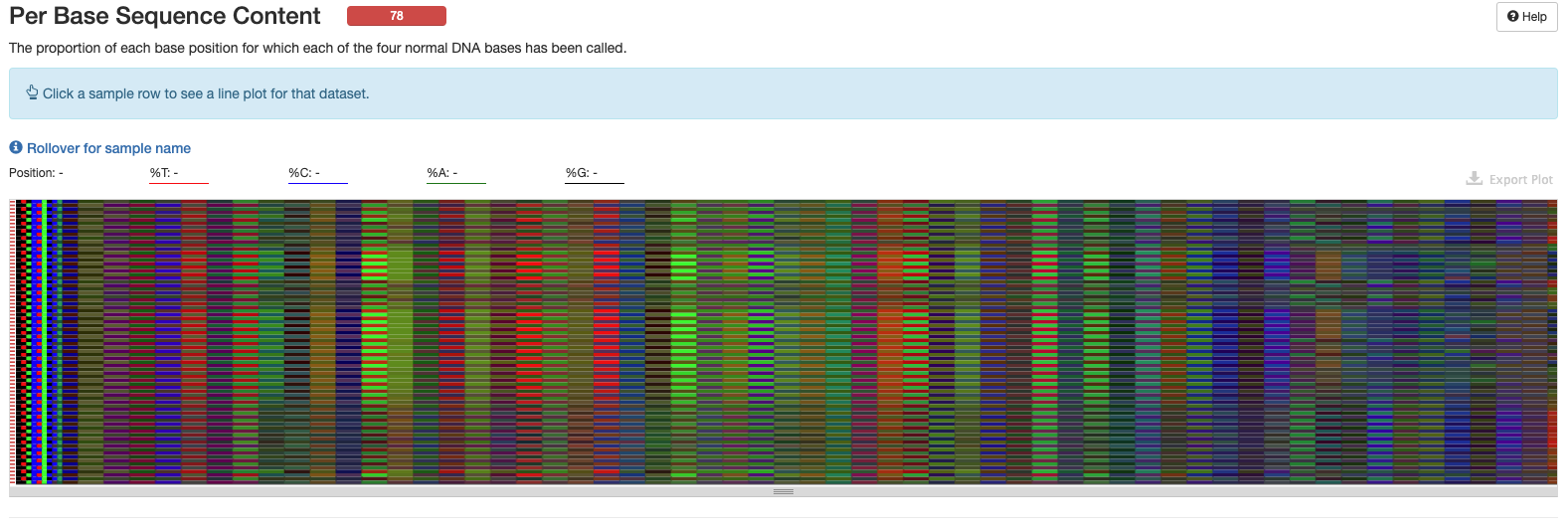
Interpretation: Amplicon libraries are expected to have bias and may not have a normal distribution. Issues at the start of eqch sequence as seen here would be expected for 16S data.
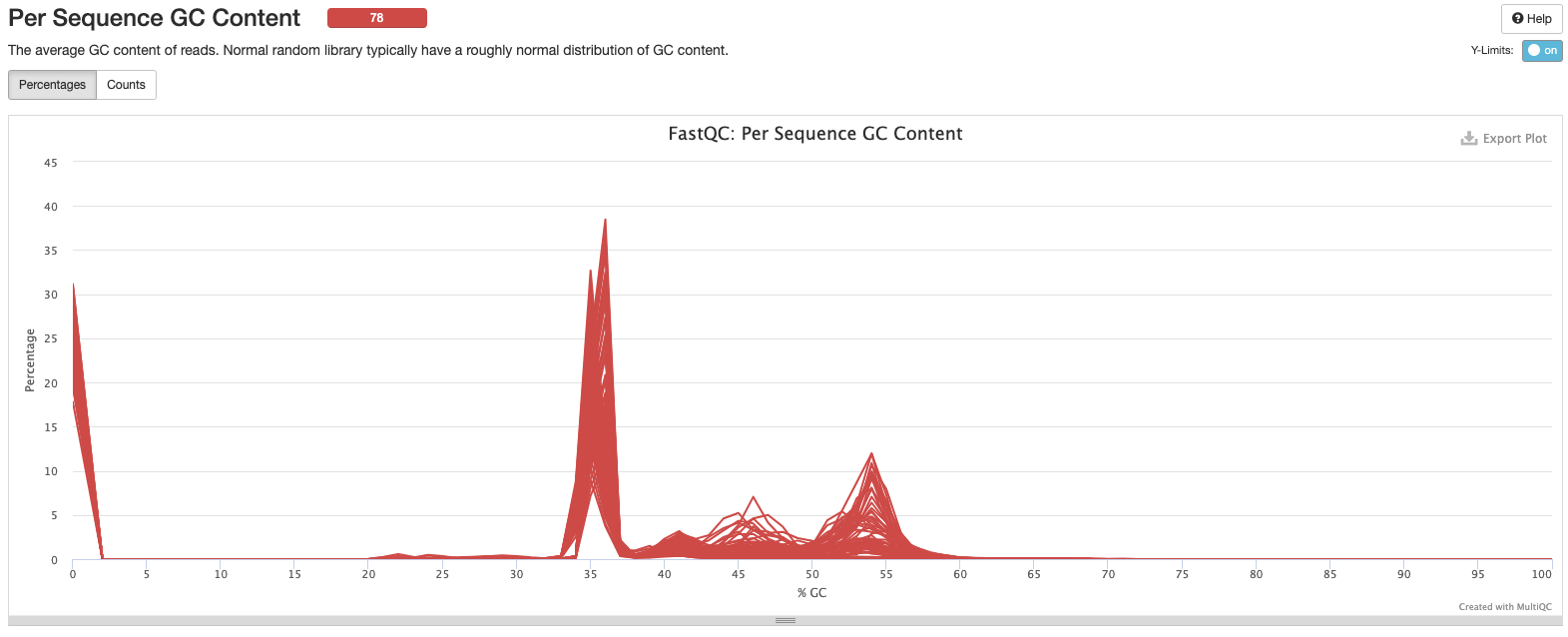
Interpretation: Peak at beginning may be resolved with trimming/cleaning. We have a double peak here at 35% GC and 55% GC. Could be due to contamination, adapters, or over represented sequences. Look into what should be expected from 16S amplicon sequencing.
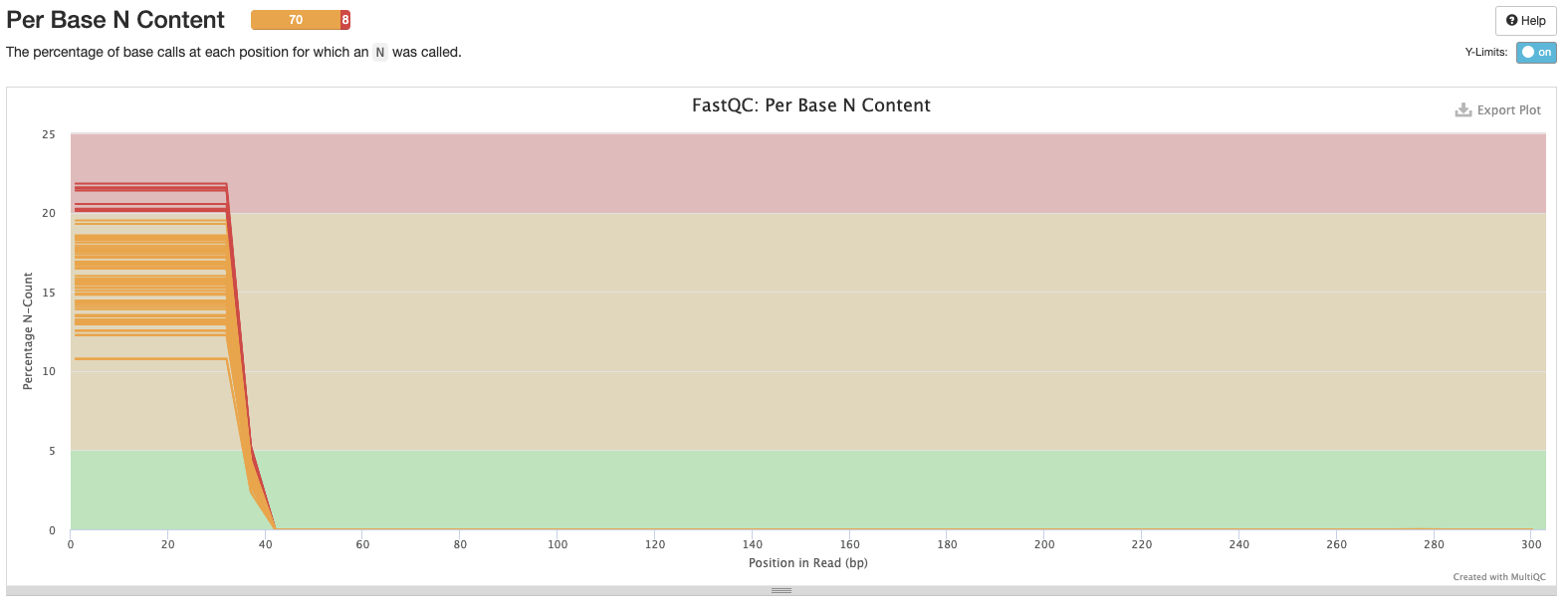
Interpretation: We can remove the problems at the start of the sequences. Low N content is as expected.
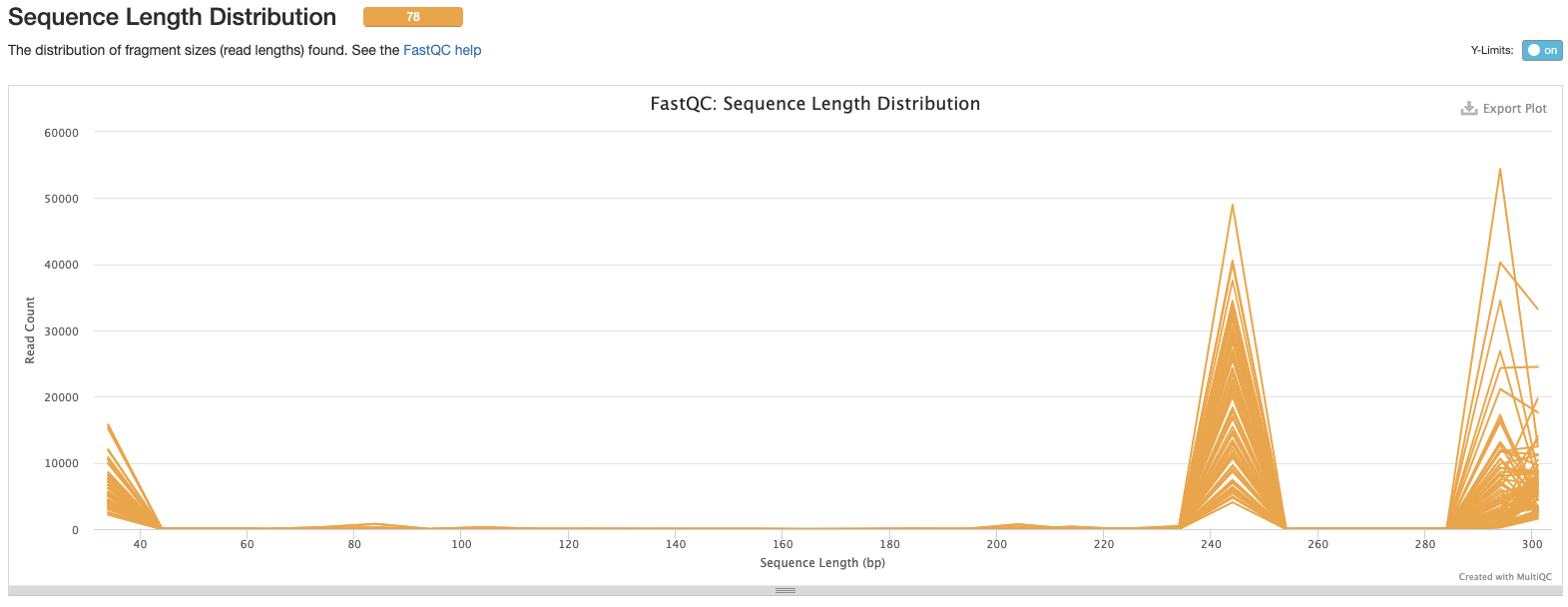
Interpretation: There is a double peak in the data at 240bp and at 280 bp. We should re evaluate this after trimming and cleaning, the lengths may be different depending on sequence quality.
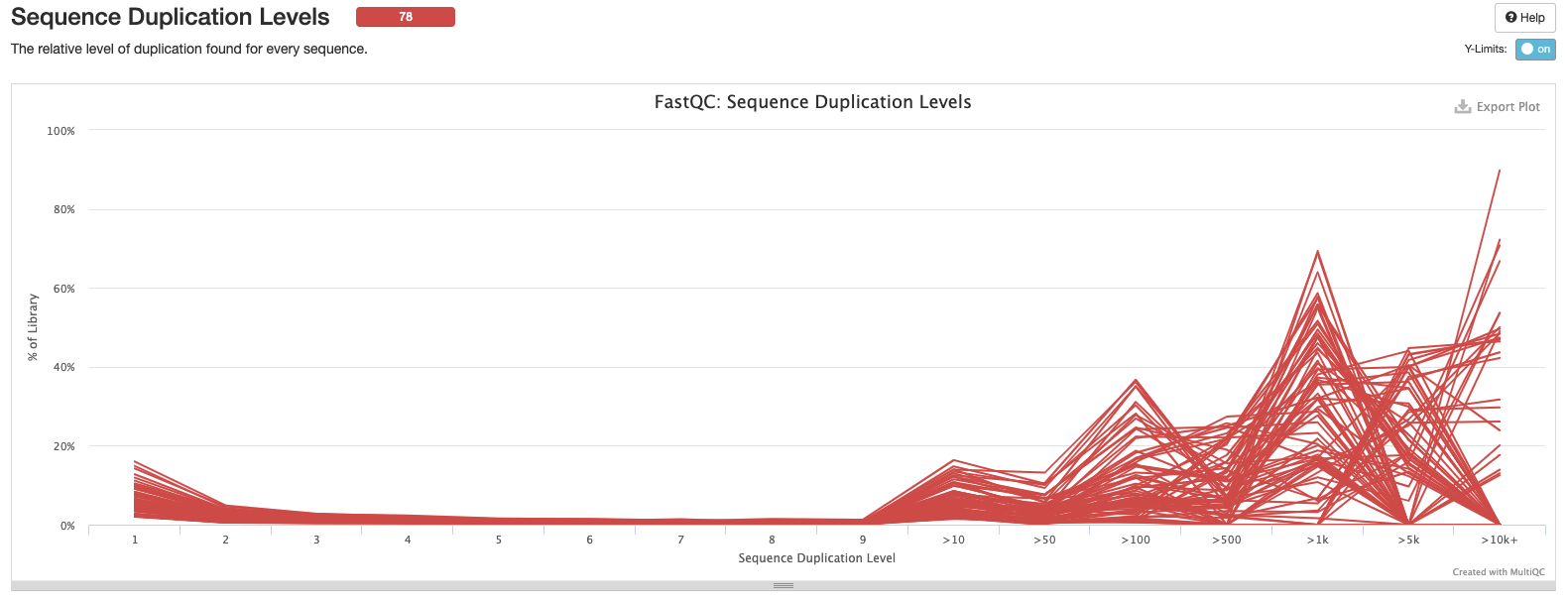
Interpretation: High level of duplication may indicate an enrichment bias in the PCR. This could mis represent the true abundance of sequences. There may also be truly over represented sequences that might be expected for microbial amplicon data.
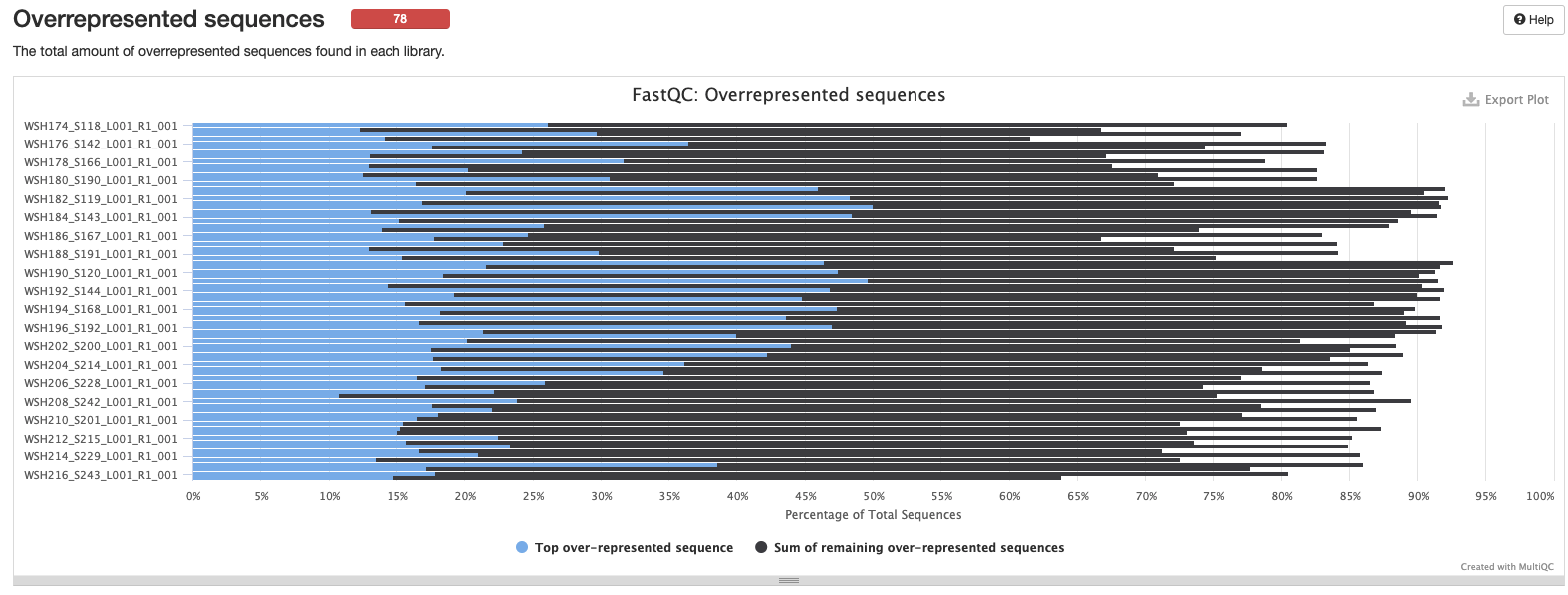
Interpretation: There are many over represnted sequences. We could blast these sequences to determine what they represent. We can also revisit this after removing adapters and other QC steps.
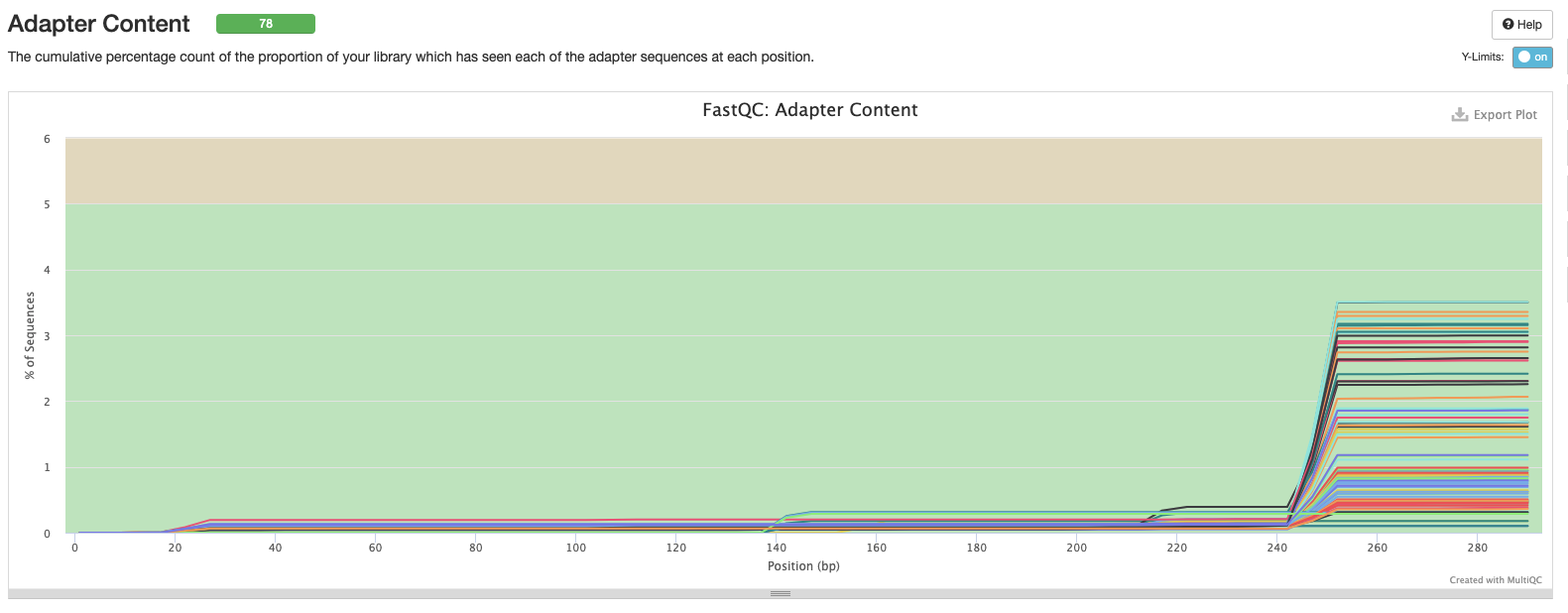
Interpretation: Adapter content can be removed with cleaning/trimming.
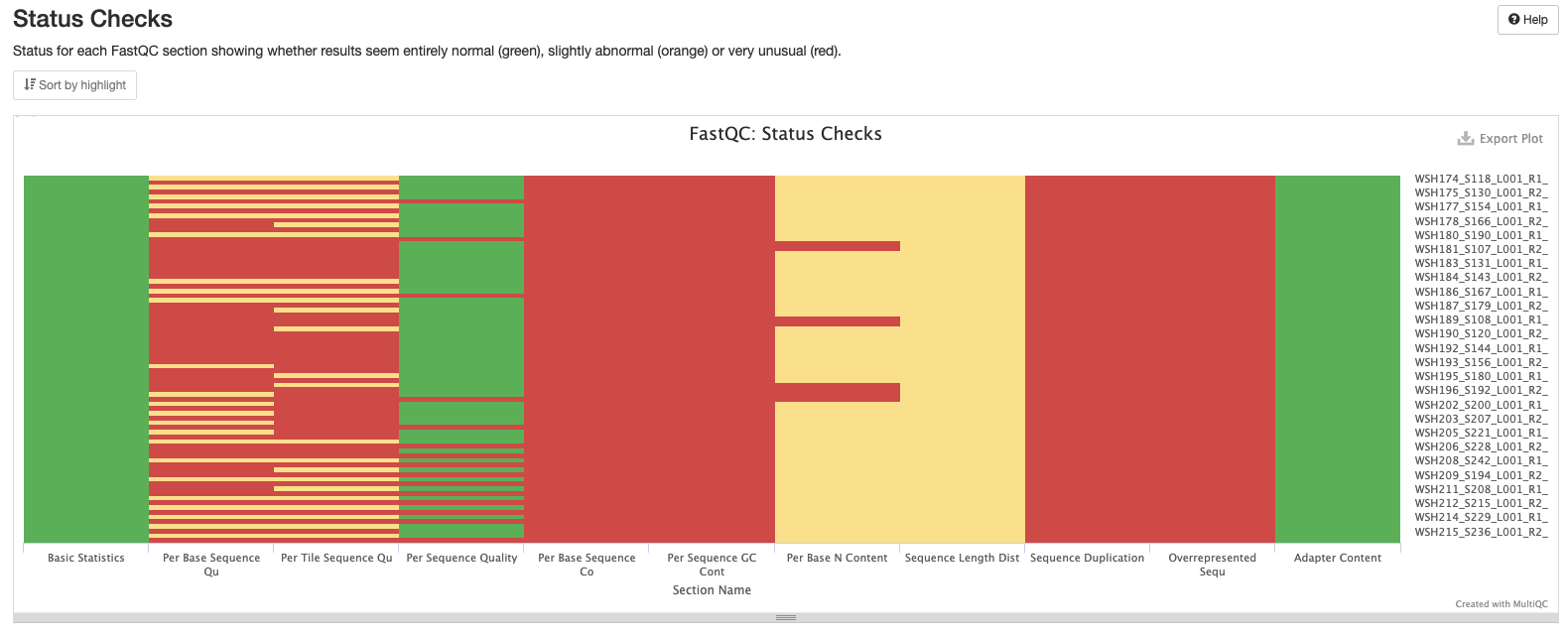
Interpretation: Shows failures and warnings as described above.


