Mcapitata Sampling and Data Collection June 2021
Post details sampling and data collection for M. capitata larval temperature exposure experiment in June 2021.
Set up and preliminary trials
6/16/2021
-
Set up prelimimary tests for respirometry equipment. Tested light levels and set up SDR in incubators according to Respirometry Protocol.
-
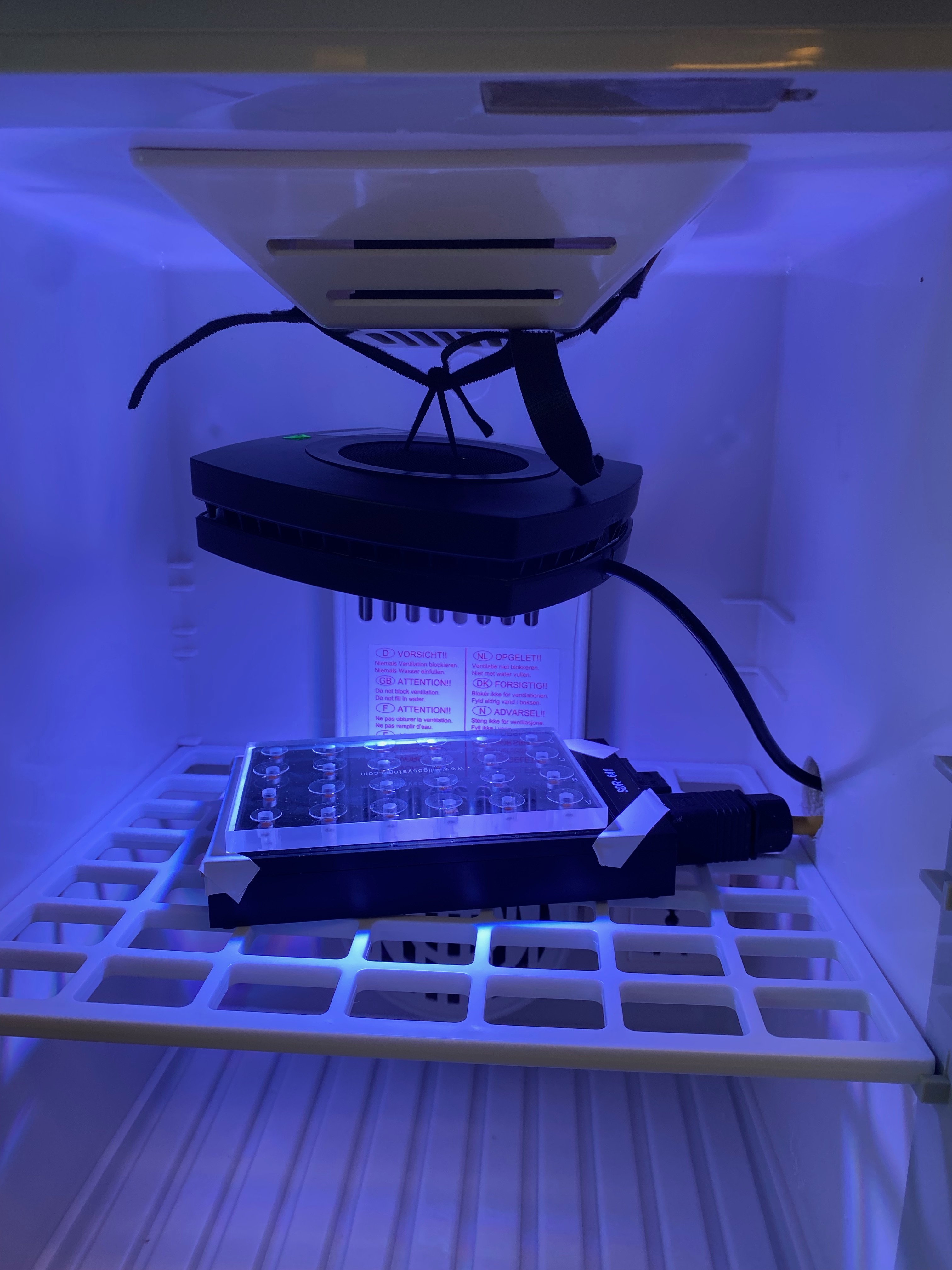
One incubator is set up at high (30C) and one at ambient (27C) with two SDRS connected to system to simultaneously conduct respirometry measurements at two temperatures. Aqua Illumination puck light added to the incubators for use in light cycles for photosynthesis measurements. SDR 641 and SDR 710 used.
-
Conducted trial run on extra larvae from buckets at 900 PAR light levels (10% intensity on all channels, no UV channel). Use 30-40 minutes in each dark and light phase for respirometry measurements. Signals obtains for 6 larvae per well. Run light and then dark to normalize light environment and avoid oxygen depletion.
- Labeling tubes, cleaning glass vials in preparation for full sampling on 6/18/2021.
Respirometry
6/18/2021
- Drained conicals and moved larvae into jars per conical (labeled A1-A6 and H1-H6) at 1100. All conicals cleaned. Left jars floating in temperature treatment until use.

- Conducted respirometry measurements as described in protocol referenced above. Ran 2 runs of respirometry at 1460 and 1800. Each run included both SDRs at 27C and 30C. Two wells with 1um Filtered Seawater were included as blanks on each SDR for each run. Six larvae were added into each well with n=2 wells per conical per run, such that all conicals were run on each SDR at each temperature (reciprocal temperature exposure). Larvae rinsed with 1um FSW prior to loading. Once all larvae were loaded, wells were filled with 1um FSW and then covered with small class cover slips. n=2 blanks per run and n=22 larval wells per run per plate. Plates were moved into incubators at 900PAR and then the light phase of oxygen measurements were conducted to capture photosynthesis. After 30-40 minutes, lights were turned off, immediately beginning the dark respiration phase. Positive and negative slopes were achieved as expected for all wells with no slope in the blanks. Metadata recorded on Drive and files downloaded onto Google Drive. Scripts, data, and analyses are on the Symbiont Integration GitHub repo.
Stable Isotope Incubations
Conducted incubations of larvae in the presence of C13 stable isotopes to allow for stable isotope metabolomic analysis of the host, will provide insights on metabolites present in host that were generated by the symbiont for nutritional exchange.
Conducted 4.5 hour incubations of coral larvae for stable isotope pulse-chase metabolomics. Following protocols from Hillyer 2017, Hillyer 2018 methods.
Made 4mM solutions of the following:
-
C13 bicarbonate in seawater = 280mL of 1um FSW + 108 mg C13 bicarbonate + dusting of NaOH to correct pH to 8.0
-
C12 bicarbonate in seawater (serves as chemistry control) = 240 mL 1uM FSW + 100 mg C12 bicarbonate + dusting NaOH, pH=8.0
-
1um FSW with no label as seawater controls and backups in case stable isotopes don’t work
Set up vials with incubation treatments for each temperature treatment (27C, 30C). In each temperature treatment:
n=6 C13, n=1 per conical n=6 C12, n=1 per conical n=6 FSW, n=1 per conical n=2 C13, n=2 from pooled larvae of all conicals, dark controls
1mL of larvae added to each vials
Caps not sealed to allow for air exchange
Lights set to 900 PAR, wrapped n=2 dark controls with foil so no light could get in, will be used as controls to determine if host uptakes C13, which it shouldn’t - C13 products should be symbiont in origin.
Vials moved into incubators.

After incubations, vials were sampled. Sampled by using separate equipment for each isotope type to avoid C13 contamination. Poured vial through cell strainer, rinsed in FSW 1uM in 6 well plates 3x, pipetted into tube. Tube then centrifuged 1.5min @ 13000, removed all water, flash froze and stored at -80C. Sampling between 0030 and 0200 on 6/19/2021, n=1 tube per vial, n=40 tubes.
Sampling
6/18/2021
-
Molecular (M) samples: Sampled 200 uL of concentrated larval pool, n=1 tube per conical, n=12 total. Flash frozen and stored at -80C May need futher water removal. Sampled at 1500.
-
Shield (S) samples: Sampled as in M tubes, but stored in 400uL DNA RNA Shield and put in 4C fridge. Sampled at 1500. 200 uL larvae, n=1 per conical, n=12 total tubes.
-
Physiology (P) samples: Sampled 100uL larvae, n=1 per conical, n=12 total, Larvae stored in 4% PFA and put at 4C. Sampled at 1545.
-
Energetics (E) samples: Sampled 200uL larvae for lipids, carbs, and protein with n=1 tube per conical per metric. n=36 total samples. Flash frozen and stored at -80C, may require more water removal. Sampled at 1445.
-
Biomass (B) samples: Sampled 200uL of larvae, n=2 per conical, n=24 total. Flash frozen, stored at -80C. Sampled at 1730, can use for biomass normalization.
-
Chlorophyll (C) samples: Sampled 200uL larvae, n=2 per conical, n=24 total. Flash frozen, stored at -80C. Sampled at 1815.
-
Extra samples: Sampled some leftover larvae at 2200, filled tubes with 1.5mL larvae, spun down, removed water, flash frozen, stored at -80C in “B” box.
Return unused larvae
Larvae not used in sampling were returned to treatment tanks, floating in jars at the correct treatment temperature at 0300 on 6/19/2021.


