Moorea 2023 Symbiotic Nutritional Exchange Project Daily Entries
This post details daily entries for the Fall 2023 Moorea Field Expedition.
The goal of this expedition is to conduct research within Ariana Huffmyer’s National Science Foundation Ocean Sciences Postdoctoral Fellowship research.
Experimental design and overview of this project including questions of interest and object can be viewed below.
The GitHub repository with all information and data for this project can be found here.
Thursday - November 2, 2023
Today we worked on finding our equipment, setting up our supplies, and making experimental plans. We started setting up our equipment for PI curves/respirometry, coral collection, and spawning.
We will be using the SMILE systems described in my past posts here for Acropora spp. spawning.
We talked with folks from CRIOBE and they observed high Acropora pulchra spawning last month. Hollie and Pierrick cracked a couple fragments of colonies at Mahana and didn’t see any eggs. We decided to not focus on A. pulchra and instead bring in some A. hyacinthus, which spawn in +8 to +10 days after the full moon. We will bring in colonies tomorrow/Saturday.
Hollie and Ariana went to Tahiti to help Nyssa Silbiger’s group with sampling on Friday.
Friday - November 3, 2023
Ariana and Hollie helped Nyssa’s group sample corals and water on Tahiti at three sites.
When we got back to Moorea Ariana labeled whirl packs for collecting adult and recruit samples for PI curves tomorrow. Ariana labeled whirl packs for the following corals:
Acropora: Adults = ACR A1-A5 Recruits = ACR R1-R10
Pocillopora: Adults = POC A1-A5 Recruits = POC R1-R10
Porites: Adults = POR A1-A5 Recruits = POR R1-R10
We prepared a hammer and chisel, knife, collection bags, and ruler with color standard for collections tomorrow.
Pierrick and Chloe collected 7 Acropora hyacinthus colonies (20-30 cm diameter) from Manava and placed them in large blue bin tanks at Gump in preparation for spawning. Pierrick also collected a set of test recruits of Pocillopora (n=8) to have as practice for PI curves and respiration measurements.
Pierrick and Chloe added tubing and other spawning equipment into a bleach bath to clean before spawning.
Saturday - November 4, 2023
Preparing spawning and setting up tanks
This morning, Pierrick and Chloe collected 6 more Acropora hyacinthus colonies (20-30 cm diameter) from Manava and added them to the blue bins (07:00-08:00). 4 corals were in each blue bin with high water flow. The electricity is not working at the tanks, so the corals didn’t have a pump last night.
Pierrick and Ariana then collect adults and recruits (see below) at Ava’iti (11:00-12:00) and collected 3 more A. hyacinthus from this site. We now have 16 total colonies.
Because the pumps were not working, Hollie and Chloe purchased a large pump at Polymat. We then moved all corals to a new large blue bin. The corals had very high water flow and water circulation after being added to the new tank at 15:00. All corals looked healthy.
While Ariana and Pierrick were collecting colonies, we saw eggs in a broken fragment. Eggs were mature - so we expect spawning to happen this month in at least some of the colonies.

We set up an adapted version of our spawning larval rearing systems at 15:00. Rather than using the coolers as a water delivery system in the SMILE systems, we put 12 squaricals on a water table and assembled a tubing manifold with 12 outlets that was tied above the table. This will allow us to supply 50 um filtered seawater from our canister filters on this line directly to the 12 squaricals. Squaricals will drain through banjo filters into the water table. We bleached and cleaned all equipment and added new hot glue seals to the banjo filter outlets.
This is adapted from last year’s spawning projects in Moorea detailed in my Moorea 2022 expedition notebook post here.
Here is a look at our initial set up.

We did not observe setting or spawning tonight.
Collecting adults and recruits for PI curves
Pierrick and Ariana collected adult and recruit samples for the Symbiotic Exchange project. We decided to sample Acropora, Pocillopora, and Porites corals from the Ava’iti site. The original experimental design only included Acropora and Pocillopora. We have the capacity to add in Porites and it is a relevant species for this study. We will adjust the experimental design as we go through the study to account for another species within the same number of total samples.
Today we collected 5 adult and 10 recruit samples from each species. We collected fragments from adult colonies. We are aiming for a genera level comparison because we will only be able to identify species in small recruits after DNA analysis. Therefore, we will collect adult samples representative of the species at our site so that the species composition of the adults and recruits are (hopefully) equally mixed. We will collect recruits across a range of sizes from ~ 1-4 cm in diameter. From intial scouting, it will be easier to find very small recruits of Pocillopora, but it will be harder to find very small recruits of the other species.
Collection protocol
Our goals for collection for each species were as follows.
Acropora: Sample recruits <5 cm, across a range of sizes from as small as possible to no more than 5 cm diameter. Sample recruits that are clearly separated from adult colonies to avoid any collection of small corals resulting from fragmentation. May include multiple Acropora species due to difficulty distinguishing species from smallest recruits. We will collect adult fragments using a chisel and recruits using a hammer and chisel.
Pocillopora: Sample recruits <5 cm, across a range of sizes from as small as possible to no more than 5 cm diameter. We should be able to find very small recruits pretty easily. May include multiple Pocillopora species due to difficulty distinguishing species from smallest recruits. We will collect adult fragments using a chisel and recruits using a dive knife or chisel.
Porites: Sample recruits <5 cm, across a range of sizes from as small as possible to no more than 5 cm diameter. Massive Porites small corals can often result from fragmentation or mortality of a larger colony leaving small sections behind. To avoid this, we will sample recruits that are far from other adult tissue. May include multiple massive Porites species due to difficulty distinguishing species from smallest recruits. We will collect adult and recruit fragments using a hammer and chisel.
Today we collected the following sample numbers:
Acropora: Adults = ACR A1-A5 Recruits = ACR R1-R10
Pocillopora: Adults = POC A1-A5 Recruits = POC R1-R10
Porites: Adults = POR A1-A5 Recruits = POR R1-R10
Here is the protocol we followed for collection today:
Materials:
- 2 mesh bags with clips
- Hammer and chisel
- Dive knife
- Labeled whirl packs
- Ruler with color scale
- Camera
Procedure:
- One person will collect the corals (“collector”) and the other will take pictures and hold the bags of collected corals (“holder”).
- Swim on the reef until finding a coral to sample using the criteria described above.
- Take out a bag corresponding to the correct life stage. Bags are labeled “species-lifestage-number”. For example, ACR-R-1 is Acropora-recruit-1. See list above for numbers sampled today.
- The collector will hold the ruler with color scale and the labeled whirlpack next to the coral. The holder will take a picture with the label and ruler included.
- The collector will then take the sample using a knife or hammer and chisel as needed. Try to take as little rock or other material as possible, but try not to break the recruit.
- The holder then puts the sample in the whirlpack, fills the whirlpack with water, secures the whirlpack, and puts it in the mesh bag.
- Repeat until all corals are sampled. Today we collected all POC, then collected ACR and POR as we came across them.
- Put the corals in a cooler of water to keep them submerged. Back at the lab, put the corals in a tank or water table. Put on plugs ASAP (see below).
- Upload photos to Google Drive (link below).


Overall, following this protocol worked very well. We were successful in collecting all samples (45 total) within about 1.5 hours. We were able to sample across a size gradient for all species. With two people the process was pretty smooth. The corals were brought back in filled whirl packs and placed in the water tables at 12:00. Bags were opened to allow for water flow while submerged until we were ready to add them onto plugs at 13:30.
This site will be good to return to for the rest of the study.
Photographs of the corals in field collections and a photograph of all plug corals are in today’s Google Drive folder here.
Adding adult and recruit samples to plugs for PI curves
After collection, we added the adult and recruit samples to plugs using the following steps.
- Prepare plugs by drying them and writing the sample ID/number on the plug stem with permanent marker.
- Add a drop of coral glue onto the plug.
- Trim off any excess material, organisms, or rock from the coral sample.
- Take the coral out of the water and dry the base with a paper towel.
- Place the coral onto the plug out of the water on a rack. Use more glue if needed.
- Leave to set for ~1-2 minutes.
- Return the plug with coral sample to the water table on a rack.
- Repeat in batches of 3-4 corals at a time.
- Maintain high water flow and circulation.
- Take an image of corals on plugs with a ruler and color scale as seen below.
We noticed that we accidentally collected 11 recruits of ACR and 4 adults. ACR-A-2 was incorrectly used to collect a recruit. We used that recruit as a test sample and took an adult fragment from one of the corals we brought to the lab from our same site. This adult is labeled ACR-A-2 and a new image was taken in the lab tank.
Here are some example images.

In this image, the first row (bottom) is a test set of POC with 1 ACR and 1 POR. The second row (second from bottom) is ACR samples, with the first 5 being adults and the last 10 being recruits. The third row (third from bottom) is POC and the fourth row (top row) is POC. These are all in the order of A-1 through A-5 followed by R1-R10 for each species.


These samples will be used for PI curves tomorrow.
Sunday - November 5, 2023
Today we prepared equipment and tests for PI curves, prepared sampling probes, and monitored for spawning.
Checking on adults and recruits
The adults and recruits on plugs looked good today at 08:00. No tissue mortality. Possibly some slight bleaching on adult POC tissues that we will keep an eye on.
Running PI curves
Here are the things we did today to prepare for running PI curves tomorrow:
- Read through protocol
- Prepped chambers, trimmed stands, trimmed and re glued corals that were too tall (POC and ACR adults). Trimmed fragments with coral saw at 16:00.
- Cleaned lab and set up area
- Set up OXY 10 units and equipment using the Putnam Lab OXY protocol.
- Calibrated 0% sodium sulfite and 100% airsaturated 50 um FSW (continuous bubbling for 30 min). We made the solutions and then calibrated the probes all in 0% and then all in 100% seawater following the protocol above. Probes were allowed to sit in the 0% solution for about 30 minutes prior to calibration. The calibration values for all probes are as follows. Calibration was conducted from 15:30-16:30. Calibration done at 21°C.
| Probe | 0% Phase | 100% Phase |
|---|---|---|
| 1 | 51.38 | 22.09 |
| 2 | 52.40 | 22.76 |
| 3 | 51.82 | 23.87 |
| 4 | 51.43 | 23.87 |
| 5 | 50.56 | 24.42 |
| 6 | 54.31 | 22.52 |
| 7 | 53.55 | 21.85 |
| 8 | 51.33 | 22.61 |
| 9 | 54.41 | 22.26 |
| 10 | 51.42 | 21.88 |
| 11 | 49.33 | 22.67 |
| 12 | 51.58 | 24.94 |
| 13 | 51.09 | 24.33 |
| 14 | 54.24 | 23.19 |
| 15 | 51.36 | 22.17 |
| 16 | 52.01 | 22.39 |
| 17 | 51.18 | 22.34 |
| 18 | 49.34 | 21.66 |
| 19 | 51.60 | 20.28 |
| 20 | 51.31 | 21.94 |
- Prepared Apex unit to control water temperature in the OXY chamber bath. Also prepared two water baths below the OXY table to pre heat temperatures for when we do multiple temperature runs for P & R rates. Used 1 Apex unit, 1 EB8 energy bar, 2 PM1s, and 3 temperature probes. Set up on Apex Fusion in the Putnam Lab account.
- Calibrated pH and conductivity probes. Calibrated pH probe with Tris batch T27. Conductivity probe calibrated with 1413 uS/cm at 20.7°C. Temperature probe is RS-232. Apogee probe is MQ-S10 SSN 3706.
| tris.date | mVTris | Ttris |
|---|---|---|
| 20231105 | -77.5 | 21.05 |
| 20231105 | -77 | 21.48 |
| 20231105 | -76.5 | 22 |
| 20231105 | -75.8 | 22.5 |
| 20231105 | -75.2 | 23 |
| 20231105 | -74.6 | 23.5 |
| 20231105 | -74 | 24 |
| 20231105 | -73.4 | 24.5 |
| 20231105 | -72.7 | 25 |
| 20231105 | -72.1 | 25.5 |
| 20231105 | -71.5 | 26.01 |
| 20231105 | -70.9 | 26.52 |
| 20231105 | -70.3 | 27.01 |
| 20231105 | -69.6 | 27.53 |
| 20231105 | -69.1 | 28.02 |
| 20231105 | -68.4 | 28.51 |
| 20231105 | -67.7 | 29 |
| 20231105 | -67.1 | 29.49 |
| 20231105 | -66.4 | 30 |
- Collected daily measurements at 15:00 in the water table (with corals on plugs) and in the blue tank (with A. hyacinthus adults). Weather was rainy and cloudy, so light is low.
| date | time | tank | pH.mV | temp.C | sal.psu | par | tris.date | notes |
|---|---|---|---|---|---|---|---|---|
| 11/5/23 | 1500 | water_table | -70.9 | 27.35 | 36.02 | 15 | 20231105 | rainy, cloudy |
| 11/5/23 | 1500 | blue_tank | -69 | 27.84 | 36.11 | 25 | 20231105 | rainy, cloudy |
- Optimized lights with 6 16HD light array and took light measurments at each position in the chambers. We tried using 2 32 HD lights but the light was too intense in the center of the array. We are using 18 chambers in this arrangement below. We used 6 AI 16HD lights and light was within 50 PAR between all 18 positions. Hollie will take full light measurements tomorrow for all positions.
Wall
| 7 | 8 | 9 | 16 | 17 | 18 |
| 4 | 5 | 6 | 13 | 14 | 15 |
| 1 | 2 | 3 | 10 | 11 | 12 |
Bench/people space
- Determined light levels to use for PI curves. We will run 3 PI curves tomorrow. Each run will have 2 blanks, 1 temperature probe within a chamber, and 15 coral samples. We have 15 samples of each species today, so we will do 3 PI curve runs tomorrow. Because we do not have any OXY temperature probes (oops!), we will deploy the temperature logger (Hobo Tidbit) logging every 1 second to match the frequency of data collection in the Oxy 10 system. We can then use this to back calculate oxygen solubility when analyzing the data. The water bath does not stay constant enough to rely only on a static temperature input.
Light levels for PI curves:
| PAR | Percent |
|---|---|
| 0 | 0 |
| 9 | 2 |
| 29 | 5 |
| 79 | 10 |
| 143 | 15 |
| 206 | 20 |
| 317 | 30 |
| 514 | 50 |
| 700 | 75 |
| 846 | 100 |
Preparing for the next sampling
Today Pierrick fixed the coral saw, which we will use for adding corals to plugs and trimming fragments as needed.
Tomorrow we will collect the following corals, which will supply about 1.5-2 days of PR runs:
- ACR-A-6 to ACR-A-17
- ACR-R-11 to ACR-R-30
- POC-A-6 to POC-A-17
- POC-R-11 to POC-R-30
- POR-A-6 to POR-A-17
- POR-R-11 to POR-R-30
96 total corals. I expect this will take about 3-4 hours to collect. The last time we collected it was half this amount collected in about 1.5 hours.
Ariana rinsed and dried a new set of plugs. Ariana also labled whirlpacks with these coral ID’s and labeled DNA tubes (yellow cryo labels with date, “DNA”, coral sample ID, and running consecutive number; tubes 1-96). Field equipment, bags, and whirlpacks were prepped. Tubes were labeled and placed in boxes.
Sample manifest and DNA lists are on GitHub.
Adjusting plans
We will revise the P & R rates protocol to run the following profile:
- Ik PAR (15 min): P rates
- 0 PAR (15 min): LEDR rates
- Ik PAR (5-10 min): light exposure for preparation for molecular sampling
- Then, sample a clipping for RNA into RNA/DNA shield
- Then, sample remaining fragment into whirl pack and flash freeze.
Spawning
No spawning observed.
Monday - November 6, 2023
Collecting adults and recruits
Today Ariana and Pierrick collected all the corals we wanted for PR measurements including:
- ACR-A6 to ACR-A17
- ACR-R11 to ACR-R30
- POC-A6 to POC-A17
- POC-R11 to POC-R30
- POR-A6 to POR-A17
- POR-R11 to POR-R30
We collected for about 4 hours between 09:00 to 13:00. We were able to find all recruits and adult samples using the protocol described on November 4. The only deviation from this protocol is that we included an additional photograph to get both width/planar photograph and height photograph to help us calculate total colony size.
During collection samples were enclosed in filled large whirlpacks and kept in a mesh bag hanging below the boat in between collections. Colonies looked healthy when we brought them back to the lab at 13:30. We put the corals in the water table and opened the bags.
Adding adults and recruits to plugs
We added corals to plugs using the protocol described on November 4. We used a coral diamond saw (Gryphon) to trim off any rock and excess area or algae/organisms in adult fragments. We added a total of 96 corals (all collected today) to labeled plugs. Plugs were placed in a plug mat in the water tables. Plugs were added from 16:00-18:30.
It was dark when we finished the plugging, so I didn’t take a picture. I will do this tomorrow morning.
Daily measurements
Chloe collected daily measurements at 12:00 today. We will take 3-4 measurements each day for the next few days to get higher frequency measurements across different times of day.
| date | time | tank | pH.mV | temp.C | sal.psu | par | tris.date | notes |
|---|---|---|---|---|---|---|---|---|
| 11/6/23 | 12:02 | water table | -69.9 | 28.36 | 35.95 | 376 | 20231105 | rainy, cloudy |
| 11/6/23 | 12:05 | blue tank | -69.3 | 28.8 | 35.95 | 517 | 20231105 | rainy, cloudy |
PI curves
Today we started running PI curves of adults and recruits of each species. The PI curves will allow us to determine light levels for upcoming P&R measurements. We ran three runs of PI curves with 2 today and 1 tomorrow. We set up two OXY 10 units with the first controlled by a PC (10 probes; channels 1-10) and the second controlled by a PC tablet (8 probes; channels 11-18).
We do not have the temperature probes. We are going to set a constant temperature with a water bath controlled by an Apex unit. We will launch a logger in the water bath to track temperature and apply calculations for oxygen solubility.
We can run 15 coral samples, 1 logger chamber, and 2 blank chambers in each run.
We have 15 corals per species, so this will equal 3 runs total. We will randomly mix species and lifestage within each run.
The PI curve protocol will follow the Putnam Lab PI protocol.
Data, metadata, and run information sheets are on GitHub here.
Data are recorded in umol/L. Temperature was set at 27.8°C. Stir bars are spinning at 380 on VELP Scientifica multistirrer digital magnetic stirrer plates. Chamber volume is recorded at the end of the run. The run information is below.
Blanks are labeled as BK-# in running order across the entire project/experiment. Blanks had just plugs. Water was loaded at treatment temperature with raw seawater.
At the end of each run, corals were put in their whirl packs and stored at -20°C. We will clip these for DNA sequencing at a later date.
Corals were exposed to 5 min of light at 50% to standardize the light environment prior to measuring oxygen consumption at 0 PAR for light enhanced dark respiration (LEDR) at the start of the run.
This is what our set up looks like.


Here is the run metadata from the 2 runs today:
| Run | Light_Level | Light_Value | Date | Start.time | Stop.time | Light_Percent |
|---|---|---|---|---|---|---|
| Run1 | 0 | 0 | 20231106 | 11:12 | 11:27 | 0 |
| Run1 | 1 | 9 | 20231106 | 11:29 | 11:44 | 2 |
| Run1 | 2 | 29 | 20231106 | 11:46 | 12:01 | 5 |
| Run1 | 3 | 79 | 20231106 | 12:05 | 12:20 | 10 |
| Run1 | 4 | 143 | 20231106 | 12:21 | 12:36 | 15 |
| Run1 | 5 | 206 | 20231106 | 12:37 | 12:52 | 20 |
| Run1 | 6 | 317 | 20231106 | 12:53 | 13:08 | 30 |
| Run1 | 7 | 514 | 20231106 | 13:10 | 13:25 | 50 |
| Run1 | 8 | 700 | 20231106 | 13:26 | 13:41 | 75 |
| Run1 | 9 | 846 | 20231106 | 13:43 | 13:58 | 100 |
| Run2 | 0 | 0 | 20231106 | 15:51 | 16:06 | 0 |
| Run2 | 1 | 9 | 20231106 | 16:08 | 16:23 | 2 |
| Run2 | 2 | 29 | 20231106 | 16:25 | 16:40 | 5 |
| Run2 | 3 | 79 | 20231106 | 16:42 | 16:57 | 10 |
| Run2 | 4 | 143 | 20231106 | 16:59 | 17:14 | 15 |
| Run2 | 5 | 206 | 20231106 | 17:16 | 17:31 | 20 |
| Run2 | 6 | 317 | 20231106 | 17:33 | 17:48 | 30 |
| Run2 | 7 | 514 | 20231106 | 17:50 | 18:05 | 50 |
| Run2 | 8 | 700 | 20231106 | 18:09 | 18:24 | 75 |
| Run2 | 9 | 846 | 20231106 | 18:28 | 18:43 | 100 |
Temperatures were: 27.65, 28.08, 27.95, 28.06, 28.25, 27.93, 27.83, 28, 27.92, 27.97 measured at the start of each light step.
Here is the sample metadata from our 2 runs today:
| Date | sample_id | Chamber.Channel | Position | Run | Species | Lifestage | Temp.Cat | Chamber.Vol.L | Logger | Notes |
|---|---|---|---|---|---|---|---|---|---|---|
| 20231106 | ACR-A1 | 1 | P1-1 | Run1 | Acropora | Adult | 27.8 | 181 | 21723472 | |
| 20231106 | ACR-R5 | 2 | P2-7 | Run1 | Acropora | Recruit | 27.8 | 177 | 21723472 | |
| 20231106 | ACR-R1 | 3 | P2-3 | Run1 | Acropora | Recruit | 27.8 | 187 | 21723472 | |
| 20231106 | POC-A5 | 4 | P2-5 | Run1 | Pocillopora | Adult | 27.8 | 176 | 21723472 | |
| 20231106 | POR-R10 | 5 | P1-3 | Run1 | Porites | Recruit | 27.8 | 185 | 21723472 | |
| 20231106 | ACR-R10 | 6 | P2-6 | Run1 | Acropora | Recruit | 27.8 | 189 | 21723472 | |
| 20231106 | POR-R7 | 7 | P1-2 | Run1 | Porites | Recruit | 27.8 | 180 | 21723472 | |
| 20231106 | BK-2 | 8 | P2-9 | Run1 | Blank | Blank | 27.8 | 191 | 21723472 | |
| 20231106 | POC-R5 | 9 | P1-4 | Run1 | Pocillopora | Recruit | 27.8 | 194 | 21723472 | |
| 20231106 | POR-A1 | 10 | P1-5 | Run1 | Porites | Adult | 27.8 | 177 | 21723472 | |
| 20231106 | POC-R8 | 11 | P1-6 | Run1 | Pocillopora | Recruit | 27.8 | 179 | 21723472 | |
| 20231106 | LOG-1 | 12 | P2-4 | Run1 | logger | logger | 27.8 | NA | 21723472 | |
| 20231106 | POC-R9 | 13 | P1-8 | Run1 | Pocillopora | Recruit | 27.8 | 186 | 21723472 | |
| 20231106 | BK-1 | 14 | P1-7 | Run1 | Blank | Blank | 27.8 | 194 | 21723472 | |
| 20231106 | ACR-A4 | 15 | P2-8 | Run1 | Acropora | Adult | 27.8 | 180 | 21723472 | |
| 20231106 | POR-R3 | 16 | P2-1 | Run1 | Porites | Recruit | 27.8 | 194 | 21723472 | |
| 20231106 | POR-A4 | 17 | P1-9 | Run1 | Porites | Adult | 27.8 | 167 | 21723472 | |
| 20231106 | POC-A1 | 18 | P2-2 | Run1 | Pocillopora | Adult | 27.8 | 184 | 21723472 | |
| 20231106 | POC-R2 | 1 | P1-1 | Run2 | Pocillopora | Recruit | 27.8 | 178 | 21723472 | |
| 20231106 | ACR-R4 | 2 | P1-2 | Run2 | Acropora | Recruit | 27.8 | 190 | 21723472 | Crab on coral |
| 20231106 | POR-A2 | 3 | P1-3 | Run2 | Porites | Adult | 27.8 | 172 | 21723472 | |
| 20231106 | BK-4 | 4 | P1-4 | Run2 | Blank | Blank | 27.8 | 181 | 21723472 | |
| 20231106 | ACR-A3 | 5 | P1-5 | Run2 | Acropora | Adult | 27.8 | 182 | 21723472 | |
| 20231106 | POR-R1 | 6 | P1-6 | Run2 | Porites | Recruit | 27.8 | 191 | 21723472 | |
| 20231106 | BK-3 | 7 | P1-7 | Run2 | Blank | Blank | 27.8 | 190 | 21723472 | |
| 20231106 | ACR-R3 | 8 | P1-8 | Run2 | Acropora | Recruit | 27.8 | 190 | 21723472 | |
| 20231106 | POC-A3 | 9 | P1-9 | Run2 | Pocillopora | Adult | 27.8 | 199 | 21723472 | |
| 20231106 | ACR-A2 | 10 | P2-1 | Run2 | Acropora | Adult | 27.8 | 188 | 21723472 | |
| 20231106 | POC-R1 | 11 | P2-2 | Run2 | Pocillopora | Recruit | 27.8 | 192 | 21723472 | |
| 20231106 | LOG-2 | 12 | P2-3 | Run2 | logger | logger | 27.8 | NA | 21723472 | |
| 20231106 | POR-R2 | 13 | P2-4 | Run2 | Porites | Recruit | 27.8 | 182 | 21723472 | |
| 20231106 | ACR-R2 | 14 | P2-5 | Run2 | Acropora | Recruit | 27.8 | 180 | 21723472 | |
| 20231106 | POC-R3 | 15 | P2-6 | Run2 | Pocillopora | Recruit | 27.8 | 195 | 21723472 | |
| 20231106 | POR-A3 | 16 | P2-7 | Run2 | Porites | Adult | 27.8 | 184 | 21723472 | |
| 20231106 | POR-R4 | 17 | P2-8 | Run2 | Porites | Recruit | 27.8 | 180 | 21723472 | |
| 20231106 | POC-A2 | 18 | P2-9 | Run2 | Pocillopora | Adult | 27.8 | 184 | 21723472 |
We will do the last run tomorrow.
Spawning
We saw a small amount of spawning from some fragments and from one small patch of one large colony. We didn’t collect any samples.
Here is a colony setting in one of the fragments from our project. This is important to consider that ACR and POC are in their reproductive seasons. This will create different energetic requirements for adults compared to recruits.

Tuesday - November 7, 2023
PI Curve runs and analysis
We did one more run of PI curves today. We followed the same protocol as yesterday. Here is our run information from today.
| Run | Light_Level | Light_Value | Date | Start.time | Stop.time | Light_Percent |
|---|---|---|---|---|---|---|
| Run3 | 0 | 0 | 20231107 | 12:38 | 12:52 | 0 |
| Run3 | 1 | 9 | 20231107 | 12:54 | 13:09 | 2 |
| Run3 | 2 | 29 | 20231107 | 13:11 | 13:26 | 5 |
| Run3 | 3 | 79 | 20231107 | 13:28 | 13:43 | 10 |
| Run3 | 4 | 143 | 20231107 | 13:45 | 14:00 | 15 |
| Run3 | 5 | 206 | 20231107 | 14:02 | 14:17 | 20 |
| Run3 | 6 | 317 | 20231107 | 14:19 | 14:39 | 30 |
| Run3 | 7 | 514 | 20231107 | 14:41 | 14:56 | 50 |
| Run3 | 8 | 700 | 20231107 | 14:58 | 15:13 | 70 |
| Run3 | 9 | 846 | 20231107 | 15:15 | 15:30 | 100 |
Temperatures were measured in the chamber and measured 28.15, 27.91, 28.21 in the last three intervals. Temperatures are on target.
| Date | sample_id | Chamber.Channel | Position | Run | Species | Lifestage | Temp.Cat | Chamber.Vol.L | Logger | Notes |
|---|---|---|---|---|---|---|---|---|---|---|
| 20231107 | POC-A5 | 1 | P1-1 | Run3 | Pocillopora | Adult | 27.8 | 0.195 | 21723472 | |
| 20231107 | ACR-R8 | 2 | P1-2 | Run3 | Acropora | Recruit | 27.8 | 0.19 | 21723472 | |
| 20231107 | POR-R6 | 3 | P1-3 | Run3 | Porites | Recruit | 27.8 | 0.196 | 21723472 | |
| 20231107 | POR-R7 | 4 | P1-4 | Run3 | Porites | Recruit | 27.8 | 0.201 | 21723472 | |
| 20231107 | ACR-R7 | 5 | P1-5 | Run3 | Acropora | Recruit | 27.8 | 0.198 | 21723472 | |
| 20231107 | ACR-A5 | 6 | P1-6 | Run3 | Acropora | Adult | 27.8 | 0.406 | 21723472 | |
| 20231107 | POR-R8 | 7 | P1-7 | Run3 | Porites | Recruit | 27.8 | 0.194 | 21723472 | |
| 20231107 | POC-A4 | 8 | P1-8 | Run3 | Pocillopora | Adult | 27.8 | 0.193 | 21723472 | |
| 20231107 | ACR-R6 | 9 | P1-9 | Run3 | Acropora | Recruit | 27.8 | 0.181 | 21723472 | |
| 20231107 | BK-5 | 10 | P2-1 | Run3 | Blank | Blank | 27.8 | 0.198 | 21723472 | |
| 20231107 | LOG-3 | 11 | P2-2 | Run3 | logger | logger | 27.8 | NA | 21723472 | |
| 20231107 | POR-R9 | 12 | P2-3 | Run3 | Porites | Recruit | 27.8 | 0.194 | 21723472 | |
| 20231107 | POR-R10 | 13 | P2-4 | Run3 | Porites | Recruit | 27.8 | 0.195 | 21723472 | |
| 20231107 | POR-R5 | 14 | P2-5 | Run3 | Porites | Recruit | 27.8 | 0.194 | 21723472 | |
| 20231107 | POC-R6 | 15 | P2-6 | Run3 | Pocillopora | Recruit | 27.8 | 0.193 | 21723472 | |
| 20231107 | POC-R4 | 16 | P2-7 | Run3 | Pocillopora | Recruit | 27.8 | 0.196 | 21723472 | |
| 20231107 | BK-6 | 17 | P2-8 | Run3 | Blank | Blank | 27.8 | 0.19 | 21723472 | |
| 20231107 | ACR-R9 | 18 | P2-9 | Run3 | Acropora | Recruit | 27.8 | 0.189 | 21723472 |
We analyzed the PI curve data. We had to troubleshoot the script because the script was loading in the time column in different time zones depending on if the data was from the PC or the tablet (channels 11-18 created errors). We manually changed the times. Ariana checked the computers and couldn’t find any settings that would change the time zone. Computers are on correct time. We will have to return to this issue to analyze P&R rates.
We also used a blackout curtain today during the dark run portions to avoid any effects of the lights or door opening.
Here are the PI curves from our 3 runs:
By individual:

By group:

Dotted lines here are recruits and solid lines are adults.
Parameter estimates:

We are missing some colonies from this data because the NLS model did not fit some of the samples well. The samples that failed the NLS model are:
- ACR-A4
- POC-A5
- POR-A5
- ACR-R6
- ACR-R8
- ACR-R9
- POC-R3
- POC-R4
- POC-R6
- POC-R9
- POC-R10
- POR-R4
- POR-R5
- POR-R6
- POR-R7
- POR-R9
- POR-R10
This is due to sensitivity of the NLS model to the model fit. We wil return to these colonies to troubleshoot. Many of the POR recruits didn’t show an asymptote in the curve, which may be the issue. This is interesting that max photosynthesis wasn’t reached.
For our next step of P&R curves, we will select light levels between 500-600 PAR to capture max photosynthesis rates without hitting photoinhibition.
Daily measurements
Chloe took daily measurements at multiple points throughout the day. Daily measurement data are on GitHub here.
Plugs from yesterday
The plugs collected from yesterday look happy!

We took a photo with a scale bar for reference for future measurements of diameter/size. The rows are organized in order of sample ID:
ACR-A6 to A17
ACR-R11 to R30
POC-A6 to A17
POC-R11 to R30
POR-A6 to A17
POR-R11 to R30
Spawning
There was a little spawning tonight again, but no new corals. We will check again tomorrow.
Planning PR rates
To measure photosynthesis (P) and respiration (R) rates, we will expose corals to the following profile:
- 15 minutes at saturating irradiance (Ik)
- 15 minutes in the dark (LEDR)
- 10 minutes at saturating irradiance (Ik) to allow for photosynthetic activity to resume prior to sampling; not used for rate measurements
- Snap freeze in liquid nitrogen for RNA/DNA
We will measure n=4 adults per species per temperature (36 total adutls) and n=6 recruits per species per temperature (54 recruits). This will divide out for us to do 6 separate runs (n=2 adults per species and n=3 recruits per species per run) with 2 runs at each temperature. 90 total corals will be sampled. We will select recruits across the size gradient for each run. This will take place with 1 run per temperature per day.
This is our current working plan!
Planning isotope metabolomics incubations
For isotoped, we will collect corals and clip DNA while adding them onto plugs. We will then do a 5 h incubation at 27, 30, or 33°C at 550 PAR. After incubations, they will be rinsed in FSW and snap frozen in liquid nitrogen. We will use 15 chambers per temperature per day, running a total of 45 corals per day. One temperature run will include 15 light C13 enriched corals:
- n=3 recruits per species = 9 per run
- n=2 adults per species = 6 per run
This would give n=4 adults per species per temperature and n=6 recruits per species per temperature.
Run over 2 days would equal 15 * 3 temps * 2 days = 90 samples
After the light incubations, we will do a control run with the following replication:
- Control Run 1: n=6 dark samples (n=1 per lifestage * species)
- Control Run 2: n=6 C12 light samples (n=1 per lifestage * species) + n=6 C12 light samples (n=1 per lifestage * species)
With controls, we will have a total of 90 + 6 + 12 = 108 samples
The controls will be run on day 3.
In total we will need:
- ACR recruits = 18 + 1 + 2 = 21
- ACR adults = 12 + 1 + 2 = 15
- POC recruits = 18 + 1 + 2 = 21
- POC adults = 12 + 1 + 2 = 15
- POR recruits = 18 + 1 + 2 = 21
- POR adults = 12 + 1 + 2 = 15
This equals 108 corals. We will do a field collection where we will collect in stages:
- First trip = POC recruits, POC adults, and ACR adults (Saturday AM); 54 corals
- Second trip = ACR recruits, POR recruits, POR adults (Saturday PM); 54 corals
A team will collect in the field and another team will clip each sample for DNA and add to plugs.
We will need to label tubes, whirl packs, and plugs for the following numbers before Saturday:
- ACR-A18 to ACR-A32
- ACR-R31 to ACR-R51
- POC-A18 to POC-A32
- POC-R31 to POC-R51
- POR-A18 to POR-A32
- POR-R31 to POR-R51
Surface area of PI curve corals
We clipped each PI curve coral sample stored at -20°C for DNA (~0.5 cm area; 5-10 polyp area) placed into RNA/DNA shield. Gloves, clippers and equipment were sterilzed between clippings with 20% bleach, 80% ethanol, and DI water. Samples were stored in tube box #1 in the Molecular Lab -40°C. Stored in 500 uL RNA DNA shield.
DNA metadata and sample list is on GitHub here.
After clipping, samples were put into cups with 40-50% bleach solution. After tissue was removed, samples were put in the drying oven at 80°C at 22:00. They will be ready for wax dipping the next morning.
Wednesday - November 8, 2023
Surface area of PI curve corals
Dried skeletons were wax dipped in the morning. Ariana ran a new standard curve, which we will use for this same batch of wax for the remainder of the project. Parrafin wax used with wax bath. Weights measured on the LTER Mettler Toledo ME303TE balance.
Wax dipping was done as describe in the E5 protocol and the data is on GitHub here. All samples were measured and 7 standards were used ranging from 1.18cm to 7.44cm diameter (wooden spheres). All samples dipped with 1 layer of wax.
Planning PR rates for tomorrow and preparing tanks
Chloe prepared the water bath for the respiration measurements, cleaned chambers, prepared water bath bins, and rinsed all previous spawning equipment.
We altered our plan for PR measurements. Here is the new plan:
- We will use the same replication as previously decided. We will run 6 runs of PR measurements with 2 runs per temperature (27, 30, 33C). Each run will contain 15 corals.
- Each run will contain n=2 adults per species and n=3 recruits per species. This will result in n=4 adults per species per temperature and n=6 recruits per species per temperature.
- Rather than running PR from the corals at ambient in short incubations, we will pre-incubate corals in water bath mesocosm tanks in the mesocosm room for the same amount of time corals will be in the isotope metabolomics incubations later in the project. This will allow us to make more direct comparisons between temperature effects on metabolomics and PR rates.
- To do this, we will fill n=1 large tank per temperature and incubate the corals prior to PR measurements. We will work on setting this up today.
Based on the PI curve results, we will aim for a light exposure of 550 PAR for incubations and PR measurements. We did light measurements at 55% and 60% light intensity in the respirometry water bath to pick the correct light intensity.
Mean light intensity across 24 spots in the water bath was 531 at 60% intensity and 480 at 55% intensity. We will use 60% light intensity.
Loggers were launched in each of the three mesocosm tanks at 16:00.
- Logger #21723473 = Tank #1 (27°C)
- Logger #21723477 = Tank #2 (30°C)
- Logger #21723476 = Tank #3 (33°C)
Here is our general set up for our tanks:
- 3 tanks with lower rack ambient (27) and upper rack 30 and 33 °C temperatures
- Each tank has apex units and probes, pumps, a hobo logger, and 500W titanium heater
- Corals 5 inches above the base of the tank on a basket elevating a plug mat/rack
- Flow was 16-17 mL per 5 sec
- Logger was in middle of tank under the basket out of direct light
- Lights were 9cm from water level
- All apex probes calibrated 20231108
- Tanks were 55 cm x 55 cm x 36 cm. The volume was 108.9 L. At the water flow rate this was a 9.45 hr turnover rate (0.192 L/min).
Light values were measured in the center and 4 quandrants around the center. Light values were the following in PAR at 60% intensity:
- Tank 1 = 426, 435, 570, 501, 587
- Tank 2 = 475, 581, 483, 493, 534
- Tank 3 = 513, 435, 550, 507, 547


We added n=2 fragments per species to one tank to test that they live in these conditions for our experiment. We added them to the tank, ramped to 33°C, and kept for 5 hours. They looked good at the end! We are good to go with running these incubations.
We will put the corals in the tanks at ambient temperature (27) and ramp them to the desired temperature treatment over 30-60 minutes. Once temperature is reached, they will be incubated in tanks for 4 hours, then the PR rates will start. Corals will be sampled after about 1 hour in the respiration chambers, giving 5 hours of total incubation time. The isotope corals will also be incubated for 5 hours so we can make more direct comparisons between measurements.
Tank maintenance and daily measurements
A Hobo Tidbit logger was launched at 08:00 in the fragment water table logging every 10 minutes (SSN 21723474).
Chloe collected daily measurements multiple times today, found on GitHub here.
Testing tank and jar incubations at high temperature
We added 6 test corals to 6 glass jars that will be used for the isotope incubations. We then ran a test incubation for 5 hours at the highest temperature, 33C, to verify that the corals wouldn’t die at this temperature. These jars are 4 oz jars with a plug stand and stir bar. This will allow for circulation during the incubations. The jars will be open to allow for light exposure and prevent metabolism of respired carbon label.
Here is what our test looked like. We used n=2 of each species for this test at 33°C.

We will construct bins to hold 15 samples at each temperature simultaneously during the isotope exposures.
At the end of the exposure, the corals looked great! They also stayed alive after the incubation for the rest of the experiment.
Thursday - November 9, 2023
PR rates and sampling
Today we ran 3 runs of P&R measurements. Here is our metadata for each run.
The general protocol is (as described above for PI curves):
- Prepare the plate map of corals in each run (n=2 adults per species, n=3 recruits per species, n=1 blank). We are only able to run 16 total samples (15 corals and 1 blank) because two sensors (5 and 8) stopped working on the Oxy-10 (channels 15 and 18).
- Add the corals to the tank corresponding to the temperature treatment. Measure temperature in these tanks every 30 minutes. Ramp using Apex to the correct temperature. Record start time.
- Rearrange position of corals at hour 2 of incubation to account for light field variability.
- 30 minutes before the run needs to be measured, prepare the water bath to the correct temperature.
- Add coral plug on to the plug stand in the acrylic chamber (200 mL chambers) with a stir bar. Add the coral and fill with treatment water in the treatment tank. Seal with the acrylic lid (tubing around edge of lid) and plug the temperature probe hole (since we do not have temperature probes).
- Remove water from the top. Put the chamber on the stir plate, which is below the water bath.
- Stir at 380.
- Add Oxy 10 oxygen probes to each chamber. Check for bubbles and reseal if necessary. Make sure crabs and other organisms are removed.
- Name all measurement channels in the Oxy 10 and start probe measurement. Set temperature to manual run temperature.
- Once all corals are in the bin, turn on lights to 60% (550 PAR). Run in the light for 15 minutes.
- Turn off lights, cover array with black out curtain. Run in the dark for 15 minutes. Record start and end times for each phase with 2 minutes between light changes to allow for acclimation.
- Finally turn the lights on again to 60%. Run in the light for 10 minutes.
- Stop probe measurement.
- Take corals out one at a time. Remove the coral and snap freeze in liquid nitrogen in whirl pack. Pour water into graduated cylinder and measure water volume. Record in metadata.
- Rinse all chambers to prepare for the next run of measurements.
Sampled corals were stored in the Biocode -40°C freezer in a mesh bag.
Here are examples of sealed chambers.

We ran a staggered start time for each run so that runs were incubating throughout the day.
Run 1: 33°C
Run 2: 27°C
Run 3: 30°C
Here is our sample metadata from all runs done today (and tomorrow). I added this post after all runs were done, so all data is below for Nov 9 and Nov 10 runs.
| Date | sample_id | Species | Lifestage | Chamber.Channel | Position | Run | Temp.Cat | Tank | Time.Loaded | Time.Temp.Reached | Time.5h | Time.Measured | Time.Sampled | Total.Incubation | Chamber.Vol.L | Notes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 20231108 | POR-A15 | Porites | Adult | 1 | P1-1 | Run1 | 33 | 3 | 7:30 | 8:30 | 13:30 | 12:30 | 13:30 | 5 | 0.172 | |
| 20231108 | POC-R19 | Pocillopora | Recruit | 2 | P2-7 | Run1 | 33 | 3 | 7:30 | 8:30 | 13:30 | 12:30 | 13:30 | 5 | 0.173 | |
| 20231108 | BK-1 | Blank | Blank | 3 | P2-3 | Run1 | 33 | 3 | 7:30 | 8:30 | 13:30 | 12:30 | 13:30 | 5 | 0.188 | |
| 20231108 | ACR-R13 | Acropora | Recruit | 4 | P2-5 | Run1 | 33 | 3 | 7:30 | 8:30 | 13:30 | 12:30 | 13:30 | 5 | 0.188 | |
| 20231108 | POC-R14 | Pocillopora | Recruit | 5 | P1-3 | Run1 | 33 | 3 | 7:30 | 8:30 | 13:30 | 12:30 | 13:30 | 5 | 0.186 | |
| 20231108 | ACR-A6 | Acropora | Adult | 6 | P2-6 | Run1 | 33 | 3 | 7:30 | 8:30 | 13:30 | 12:30 | 13:30 | 5 | 0.188 | |
| 20231108 | POR-R28 | Porites | Recruit | 7 | P1-2 | Run1 | 33 | 3 | 7:30 | 8:30 | 13:30 | 12:30 | 13:30 | 5 | 0.192 | |
| 20231108 | POC-A11 | Pocillopora | Adult | 8 | P2-9 | Run1 | 33 | 3 | 7:30 | 8:30 | 13:30 | 12:30 | 13:30 | 5 | 0.19 | |
| 20231108 | POR-R19 | Porites | Recruit | 9 | P1-4 | Run1 | 33 | 3 | 7:30 | 8:30 | 13:30 | 12:30 | 13:30 | 5 | 0.186 | |
| 20231108 | ACR-R16 | Acropora | Recruit | 10 | P1-5 | Run1 | 33 | 3 | 7:30 | 8:30 | 13:30 | 12:30 | 13:30 | 5 | 0.196 | |
| 20231108 | POR-R13 | Porites | Recruit | 11 | P1-6 | Run1 | 33 | 3 | 7:30 | 8:30 | 13:30 | 12:30 | 13:30 | 5 | 0.194 | |
| 20231108 | POR-A10 | Porites | Adult | 12 | P2-4 | Run1 | 33 | 3 | 7:30 | 8:30 | 13:30 | 12:30 | 13:30 | 5 | 0.194 | |
| 20231108 | ACR-A9 | Acropora | Adult | 13 | P1-8 | Run1 | 33 | 3 | 7:30 | 8:30 | 13:30 | 12:30 | 13:30 | 5 | 0.191 | |
| 20231108 | POC-A12 | Pocillopora | Adult | 14 | P1-7 | Run1 | 33 | 3 | 7:30 | 8:30 | 13:30 | 12:30 | 13:30 | 5 | 0.187 | |
| 20231108 | POC-R30 | Pocillopora | Recruit | 16 | P2-1 | Run1 | 33 | 3 | 7:30 | 8:30 | 13:30 | 12:30 | 13:30 | 5 | 0.192 | |
| 20231108 | ACR-R12 | Acropora | Recruit | 17 | P1-9 | Run1 | 33 | 3 | 7:30 | 8:30 | 13:30 | 12:30 | 13:30 | 5 | 0.182 | |
| 20231108 | ACR-R27 | Acropora | Recruit | 1 | P1-1 | Run2 | 27 | 1 | 10:30 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.19 | Given 30 minutes at 27 - no ramp needed, but kept total exposure time the same as the other treatments |
| 20231108 | ACR-A16 | Acropora | Adult | 2 | P1-2 | Run2 | 27 | 1 | 10:30 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.192 | |
| 20231108 | POC-A9 | Pocillopora | Adult | 3 | P1-3 | Run2 | 27 | 1 | 10:30 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.189 | |
| 20231108 | POR-R30 | Porites | Recruit | 4 | P1-4 | Run2 | 27 | 1 | 10:30 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.186 | |
| 20231108 | ACR-R23 | Acropora | Recruit | 5 | P1-5 | Run2 | 27 | 1 | 10:30 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.172 | |
| 20231108 | POC-R29 | Pocillopora | Recruit | 6 | P1-6 | Run2 | 27 | 1 | 10:30 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.179 | |
| 20231108 | POR-A12 | Porites | Adult | 7 | P1-7 | Run2 | 27 | 1 | 10:30 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.178 | |
| 20231108 | ACR-R11 | Acropora | Recruit | 8 | P1-8 | Run2 | 27 | 1 | 10:30 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.192 | |
| 20231108 | POR-R22 | Porites | Recruit | 9 | P1-9 | Run2 | 27 | 1 | 10:30 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.188 | |
| 20231108 | ACR-A14 | Acropora | Adult | 10 | P2-1 | Run2 | 27 | 1 | 10:30 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.193 | |
| 20231108 | POC-A6 | Pocillopora | Adult | 11 | P2-2 | Run2 | 27 | 1 | 10:30 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.189 | |
| 20231108 | POC-R11 | Pocillopora | Recruit | 12 | P2-3 | Run2 | 27 | 1 | 10:30 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.192 | |
| 20231108 | BK-2 | Blank | Blank | 13 | P2-4 | Run2 | 27 | 1 | 10:30 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.19 | |
| 20231108 | POC-R23 | Pocillopora | Recruit | 14 | P2-5 | Run2 | 27 | 1 | 10:30 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.195 | |
| 20231108 | POR-A16 | Porites | Adult | 16 | P2-7 | Run2 | 27 | 1 | 10:30 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.19 | |
| 20231108 | POR-R18 | Porites | Recruit | 17 | P2-8 | Run2 | 27 | 1 | 10:30 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.192 | |
| 20231108 | ACR-A10 | Acropora | Adult | 1 | P1-1 | Run3 | 30 | 2 | 13:00 | 13:30 | 18:30 | 17:30 | 18:30 | 5 | 0.19 | |
| 20231108 | POC-R22 | Pocillopora | Recruit | 2 | P1-2 | Run3 | 30 | 2 | 13:00 | 13:30 | 18:30 | 17:30 | 18:30 | 5 | 0.194 | |
| 20231108 | POR-R16 | Porites | Recruit | 3 | P1-3 | Run3 | 30 | 2 | 13:00 | 13:30 | 18:30 | 17:30 | 18:30 | 5 | 0.191 | |
| 20231108 | POC-A17 | Pocillopora | Adult | 4 | P1-4 | Run3 | 30 | 2 | 13:00 | 13:30 | 18:30 | 17:30 | 18:30 | 5 | 0.192 | |
| 20231108 | POR-R14 | Porites | Recruit | 5 | P1-5 | Run3 | 30 | 2 | 13:00 | 13:30 | 18:30 | 17:30 | 18:30 | 5 | 0.184 | |
| 20231108 | ACR-R26 | Acropora | Recruit | 6 | P1-6 | Run3 | 30 | 2 | 13:00 | 13:30 | 18:30 | 17:30 | 18:30 | 5 | 0.186 | |
| 20231108 | POR-A9 | Porites | Adult | 7 | P1-7 | Run3 | 30 | 2 | 13:00 | 13:30 | 18:30 | 17:30 | 18:30 | 5 | 0.188 | |
| 20231108 | POR-R29 | Porites | Recruit | 8 | P1-8 | Run3 | 30 | 2 | 13:00 | 13:30 | 18:30 | 17:30 | 18:30 | 5 | 0.194 | |
| 20231108 | POC-R26 | Pocillopora | Recruit | 9 | P1-9 | Run3 | 30 | 2 | 13:00 | 13:30 | 18:30 | 17:30 | 18:30 | 5 | 0.188 | |
| 20231108 | POC-A14 | Pocillopora | Adult | 10 | P2-1 | Run3 | 30 | 2 | 13:00 | 13:30 | 18:30 | 17:30 | 18:30 | 5 | 0.187 | |
| 20231108 | ACR-R18 | Acropora | Recruit | 11 | P2-2 | Run3 | 30 | 2 | 13:00 | 13:30 | 18:30 | 17:30 | 18:30 | 5 | 0.182 | |
| 20231108 | ACR-R14 | Acropora | Recruit | 12 | P2-3 | Run3 | 30 | 2 | 13:00 | 13:30 | 18:30 | 17:30 | 18:30 | 5 | 0.186 | |
| 20231108 | ACR-A15 | Acropora | Adult | 13 | P2-4 | Run3 | 30 | 2 | 13:00 | 13:30 | 18:30 | 17:30 | 18:30 | 5 | 0.187 | |
| 20231108 | BK-3 | Blank | Blank | 14 | P2-5 | Run3 | 30 | 2 | 13:00 | 13:30 | 18:30 | 17:30 | 18:30 | 5 | 0.179 | |
| 20231108 | POC-R13 | Pocillopora | Recruit | 16 | P2-7 | Run3 | 30 | 2 | 13:00 | 13:30 | 18:30 | 17:30 | 18:30 | 5 | 0.194 | |
| 20231108 | POR-A7 | Porites | Adult | 17 | P2-8 | Run3 | 30 | 2 | 13:00 | 13:30 | 18:30 | 17:30 | 18:30 | 5 | 0.186 | |
| 20231109 | POC-A7 | Pocillopora | Adult | 1 | P1-1 | Run4 | 30 | 2 | 7:45 | 8:15 | 13:15 | 12:15 | 13:15 | 5 | 0.186 | |
| 20231109 | POC-R24 | Pocillopora | Recruit | 2 | P2-7 | Run4 | 30 | 2 | 7:45 | 8:15 | 13:15 | 12:15 | 13:15 | 5 | 0.194 | |
| 20231109 | POR-A17 | Porites | Adult | 3 | P2-3 | Run4 | 30 | 2 | 7:45 | 8:15 | 13:15 | 12:15 | 13:15 | 5 | 0.185 | |
| 20231109 | ACR-R22 | Acropora | Recruit | 4 | P2-5 | Run4 | 30 | 2 | 7:45 | 8:15 | 13:15 | 12:15 | 13:15 | 5 | 0.188 | |
| 20231109 | POR-R21 | Porites | Recruit | 5 | P1-3 | Run4 | 30 | 2 | 7:45 | 8:15 | 13:15 | 12:15 | 13:15 | 5 | 0.188 | |
| 20231109 | BK-4 | Blank | Blank | 6 | P2-6 | Run4 | 30 | 2 | 7:45 | 8:15 | 13:15 | 12:15 | 13:15 | 5 | 0.189 | |
| 20231109 | ACR-R25 | Acropora | Recruit | 7 | P1-2 | Run4 | 30 | 2 | 7:45 | 8:15 | 13:15 | 12:15 | 13:15 | 5 | 0.187 | |
| 20231109 | POC-R18 | Pocillopora | Recruit | 8 | P2-9 | Run4 | 30 | 2 | 7:45 | 8:15 | 13:15 | 12:15 | 13:15 | 5 | 0.188 | |
| 20231109 | POC-A10 | Pocillopora | Adult | 9 | P1-4 | Run4 | 30 | 2 | 7:45 | 8:15 | 13:15 | 12:15 | 13:15 | 5 | 0.187 | |
| 20231109 | ACR-A11 | Acropora | Adult | 10 | P1-5 | Run4 | 30 | 2 | 7:45 | 8:15 | 13:15 | 12:15 | 13:15 | 5 | 0.194 | |
| 20231109 | POR-R11 | Porites | Recruit | 11 | P1-6 | Run4 | 30 | 2 | 7:45 | 8:15 | 13:15 | 12:15 | 13:15 | 5 | 0.189 | |
| 20231109 | ACR-A7 | Acropora | Adult | 12 | P2-4 | Run4 | 30 | 2 | 7:45 | 8:15 | 13:15 | 12:15 | 13:15 | 5 | 0.189 | |
| 20231109 | POC-R12 | Pocillopora | Recruit | 13 | P1-8 | Run4 | 30 | 2 | 7:45 | 8:15 | 13:15 | 12:15 | 13:15 | 5 | 0.193 | |
| 20231109 | POR-R25 | Porites | Recruit | 14 | P1-7 | Run4 | 30 | 2 | 7:45 | 8:15 | 13:15 | 12:15 | 13:15 | 5 | 0.192 | |
| 20231109 | ACR-R28 | Acropora | Recruit | 16 | P2-1 | Run4 | 30 | 2 | 7:45 | 8:15 | 13:15 | 12:15 | 13:15 | 5 | 0.191 | |
| 20231109 | POR-A8 | Porites | Adult | 17 | P1-9 | Run4 | 30 | 2 | 7:45 | 8:15 | 13:15 | 12:15 | 13:15 | 5 | 0.19 | |
| 20231109 | ACR-R19 | Acropora | Recruit | 1 | P1-1 | Run5 | 33 | 3 | 10:00 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.19 | |
| 20231109 | ACR-A17 | Acropora | Adult | 2 | P1-2 | Run5 | 33 | 3 | 10:00 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.192 | |
| 20231109 | ACR-R30 | Acropora | Recruit | 3 | P1-3 | Run5 | 33 | 3 | 10:00 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.183 | |
| 20231109 | POC-A16 | Pocillopora | Adult | 4 | P1-4 | Run5 | 33 | 3 | 10:00 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.193 | |
| 20231109 | BK-5 | Blank | Blank | 5 | P1-5 | Run5 | 33 | 3 | 10:00 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.188 | |
| 20231109 | POR-A11 | Porites | Adult | 6 | P1-6 | Run5 | 33 | 3 | 10:00 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.184 | |
| 20231109 | ACR-A13 | Acropora | Adult | 7 | P1-7 | Run5 | 33 | 3 | 10:00 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.193 | |
| 20231109 | POC-R15 | Pocillopora | Recruit | 8 | P1-8 | Run5 | 33 | 3 | 10:00 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.188 | Make sure the name of the dataframe is POC-R15 not A15 |
| 20231109 | POR-R27 | Porites | Recruit | 9 | P1-9 | Run5 | 33 | 3 | 10:00 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.186 | |
| 20231109 | POC-A8 | Pocillopora | Adult | 10 | P2-1 | Run5 | 33 | 3 | 10:00 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.183 | |
| 20231109 | POR-A6 | Porites | Adult | 11 | P2-2 | Run5 | 33 | 3 | 10:00 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.186 | |
| 20231109 | ACR-R15 | Acropora | Recruit | 12 | P2-3 | Run5 | 33 | 3 | 10:00 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.187 | |
| 20231109 | POC-R20 | Pocillopora | Recruit | 13 | P2-4 | Run5 | 33 | 3 | 10:00 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.191 | |
| 20231109 | POR-R15 | Porites | Recruit | 14 | P2-5 | Run5 | 33 | 3 | 10:00 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.189 | |
| 20231109 | POR-R24 | Porites | Recruit | 16 | P2-7 | Run5 | 33 | 3 | 10:00 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.187 | |
| 20231109 | POC-R16 | Pocillopora | Recruit | 17 | P2-8 | Run5 | 33 | 3 | 10:00 | 11:00 | 16:00 | 15:00 | 16:00 | 5 | 0.191 | |
| 20231109 | POR-A14 | Porites | Adult | 1 | P1-1 | Run6 | 27 | 1 | 12:30 | 13:00 | 18:00 | 17:00 | 18:00 | 5 | 0.192 | |
| 20231109 | ACR-R24 | Acropora | Recruit | 2 | P1-2 | Run6 | 27 | 1 | 12:30 | 13:00 | 18:00 | 17:00 | 18:00 | 5 | 0.186 | |
| 20231109 | POR-R20 | Porites | Recruit | 3 | P1-3 | Run6 | 27 | 1 | 12:30 | 13:00 | 18:00 | 17:00 | 18:00 | 5 | 0.177 | |
| 20231109 | POR-R17 | Porites | Recruit | 4 | P1-4 | Run6 | 27 | 1 | 12:30 | 13:00 | 18:00 | 17:00 | 18:00 | 5 | 0.192 | |
| 20231109 | ACR-A12 | Acropora | Adult | 5 | P1-5 | Run6 | 27 | 1 | 12:30 | 13:00 | 18:00 | 17:00 | 18:00 | 5 | 0.19 | |
| 20231109 | POC-R27 | Pocillopora | Recruit | 6 | P1-6 | Run6 | 27 | 1 | 12:30 | 13:00 | 18:00 | 17:00 | 18:00 | 5 | 0.186 | |
| 20231109 | POR-R26 | Porites | Recruit | 7 | P1-7 | Run6 | 27 | 1 | 12:30 | 13:00 | 18:00 | 17:00 | 18:00 | 5 | 0.193 | |
| 20231109 | ACR-A8 | Acropora | Adult | 8 | P1-8 | Run6 | 27 | 1 | 12:30 | 13:00 | 18:00 | 17:00 | 18:00 | 5 | 0.191 | |
| 20231109 | POR-A13 | Porites | Adult | 9 | P1-9 | Run6 | 27 | 1 | 12:30 | 13:00 | 18:00 | 17:00 | 18:00 | 5 | 0.186 | |
| 20231109 | POC-R28 | Pocillopora | Recruit | 10 | P2-1 | Run6 | 27 | 1 | 12:30 | 13:00 | 18:00 | 17:00 | 18:00 | 5 | 0.19 | |
| 20231109 | BK-6 | Blank | Blank | 11 | P2-2 | Run6 | 27 | 1 | 12:30 | 13:00 | 18:00 | 17:00 | 18:00 | 5 | 0.184 | |
| 20231109 | POC-A13 | Pocillopora | Adult | 12 | P2-3 | Run6 | 27 | 1 | 12:30 | 13:00 | 18:00 | 17:00 | 18:00 | 5 | 0.192 | |
| 20231109 | ACR-R17 | Acropora | Recruit | 13 | P2-4 | Run6 | 27 | 1 | 12:30 | 13:00 | 18:00 | 17:00 | 18:00 | 5 | 0.183 | |
| 20231109 | ACR-R29 | Acropora | Recruit | 14 | P2-5 | Run6 | 27 | 1 | 12:30 | 13:00 | 18:00 | 17:00 | 18:00 | 5 | 0.188 | |
| 20231109 | POC-A15 | Pocillopora | Adult | 16 | P2-7 | Run6 | 27 | 1 | 12:30 | 13:00 | 18:00 | 17:00 | 18:00 | 5 | 0.192 | |
| 20231109 | POC-R21 | Pocillopora | Recruit | 17 | P2-8 | Run6 | 27 | 1 | 12:30 | 13:00 | 18:00 | 17:00 | 18:00 | 5 | 0.19 |
And here is our run metadata.
| Run | Temp.Cat | Tank | Light_Level | Light_Value | Date | Start.time | Stop.time | Light_Percent |
|---|---|---|---|---|---|---|---|---|
| Run1 | 33 | 3 | 1 | 550 | 20231108 | 12:36 | 12:51 | 60 |
| Run1 | 33 | 3 | 2 | 0 | 20231108 | 12:53 | 13:08 | 0 |
| Run1 | 33 | 3 | 3 | 550 | 20231108 | 13:10 | 13:20 | 60 |
| Run2 | 27 | 1 | 1 | 550 | 20231108 | 15:09 | 15:24 | 60 |
| Run2 | 27 | 1 | 2 | 0 | 20231108 | 15:26 | 15:41 | 0 |
| Run2 | 27 | 1 | 3 | 550 | 20231108 | 15:43 | 15:53 | 60 |
| Run3 | 30 | 2 | 1 | 550 | 20231108 | 17:32 | 17:43 | 60 |
| Run3 | 30 | 2 | 2 | 0 | 20231108 | 17:45 | 17:58 | 0 |
| Run3 | 30 | 2 | 3 | 550 | 20231108 | 18:00 | 18:10 | 60 |
| Run4 | 30 | 2 | 1 | 550 | 20231109 | 12:13 | 12:28 | 60 |
| Run4 | 30 | 2 | 2 | 0 | 20231109 | 12:35 | 12:50 | 0 |
| Run4 | 30 | 2 | 3 | 550 | 20231109 | 12:52 | 13:02 | 60 |
| Run5 | 33 | 3 | 1 | 550 | 20231109 | 15:11 | 15:26 | 60 |
| Run5 | 33 | 3 | 2 | 0 | 20231109 | 15:28 | 15:43 | 0 |
| Run5 | 33 | 3 | 3 | 550 | 20231109 | 15:45 | 15:55 | 60 |
| Run6 | 27 | 1 | 1 | 550 | 20231109 | 17:03 | 17:18 | 60 |
| Run6 | 27 | 1 | 2 | 0 | 20231109 | 17:20 | 17:35 | 0 |
| Run6 | 27 | 1 | 3 | 550 | 20231109 | 17:37 | 17:47 | 60 |
All data were exported and saved on GitHub here.
Daily measurements
Chloe took daily measurements and they are on GitHub here. Logger files are also downloaded here.
Spawning
We had a big spawning night! 8-10 of our A. hyacinthus colonies spawned. We allowed the colonies to spawn in the tank all together. We then gathered the bundles from the surface using mesh bottom scoops/containers.
We then pooled the bundles into buckets to allow them to mix.
We then allocated bundles to falcon tubes and buckets to fertilize. These protocols follow previous spawning protocols developed for Montipora capitata and Acropora pulchra.
Briefly, 5 mL of bundles were added to each of 12 falcon tubes and filled to 45 mL with 50 µm FSW. Falcon tubes were placed on their side and gently rocked until all bundles were broken and eggs could fertilize (~1 h).
Remaining bundles (we had a lot!) were pooled in buckets. Once bundles separate, sperm is released. After 1 hour of pooled fertilization, the sperm water was removed using a pipette from the falcon tubes and the eggs were washed with clean water. The same proceedure was used for the buckets but rinsing eggs with a seive.
Eggs were then added to squarical containers (described above) at 1 falcon tube per squarical and to 5 large bins/coolers that will be filled stagnant water.
All eggs were added to tanks at approximately 23:30. We added water flow at ~8-10 mL per 5 seconds and left them for the night. Coolers and bins were left open (no water flow) to allow wind to gently mix and move the bundles.
We will check them in the morning!





Friday - November 10, 2023
PR rates and sampling
We completed runs 4, 5, and 6 today. See the entry from yesterday for all the data. Everything ran smoothly! Samples were stored in the Biocode -40°C freezer.
Daily measurements
Daily measurements were completed in all incubation tanks and all outdoor tanks and are recorded here.
Spawning and larval rearing
The embryos looked great today! There was minimal mortality. We cleaned the inline filters, stirred the embryos, and removed any dirty/dead material.
We did a water change on the coolers at 2200. There was no spawning tonight.
Preparing for field sampling
We prepared materials and lists for field sampling for our next round of collections, which will occur tomorrow. We will sample the following corals for our isotope experiment.
ACR-A18 to A32
ACR-R31 to R51
POC-A18 to A32
POC-R31 to R51
POR-A18 to A32
POR-R31 to R51
We labeled tubes (for DNA clipping), plugs, and whirlpacks for tomorrow.
Saturday - November 11, 2023
Sample collection
Today we sampled corals for isotope incubations, which will occur over the next three days. We followed sampling protocols described from November 6 collections.
We sampled about half on a morning trip, about half on an afternoon trip, and have 10 more left to sample in the morning that we didn’t finish.
When the corals came back to the lab, we clipped for DNA (as described above from the PI curve corals) and added samples to plugs (as described from November 6th.
Here is the DNA sample metadata and collection metadata.
Daily measurements
Daily measurements were taken at 1800. The temperature in the blue tank was a bit high (29°C), so we increased the water flow. The adult A. hyacinthus colonies will be put back on the reef tomorrow.
Acropora spawning and larval rearing
Hollie cleaned the banjo filters and periodically stirred and checked the larvae. Everything looks great! The larvae were ciliated and moving around a bit by the evening.
No spawning tonight.
Sunday - November 12, 2023
Pocillopora spawning
We saw Pocillopora spawning this morning! Details are recorded in Hollie and Pierrick’s RAPID notebooks. These larvae will be used for this study for metabolic rate and isotopic incubations (described in future days).
Sample collection
We finished sample collection for isotopes in the morning right after observing Pocillopora spawning. Pocillopora spawning and larval rearing are recorded in Hollie and Pierrick’s notebooks for the RAPID project.
Once brought back to the lab, samples were clipped for DNA as done yesterday. Here is the DNA sample metadata and collection metadata. All samples added to Box 3. We will not be using these samples in the incubations today to allow them the same recovery time as the previously collected samples. Samples were added to plugs as done yesterday.
All photos of the collections were labeled with the sample ID and stored on Google Drive.
Here are all of the metabolomics fragments.
Isotopes day 1
Protocol and notes about using C13
Today we will start running isotope incubations! The general steps are as follows:
- Mix seawater isotope solutions
- Load samples into jars
- Start incubation in water baths
- Sample at the end of incubation (5-5.5 hrs) by taking each sample out of the jar, putting it into a labeled whirlpack, and snap freezing.
- Store samples at -40°C
Because we are enriching seawater with C13 labeled sodium bicarbonate, it is critical that the isotope is contained and does not contaminate equipment, surfaces, or water that flows back to the natural environment. This is to prevent any change in the natural C13 signal in the local habitat. Here are the precautions and steps taken to prevent any C13 exposure.
- Gloves are worn at all times. Gloves are changed after handling of any equipment or samples that touched C13 water. Researchers did not touch anything with C13 contaminated gloves.
- Countertops or any surfaces were covered with bench paper and plastic coverings, which were immediately disposed of after the C13 experiments.
- Any equipment, tools, jars, or bins that touched C13 water were cleaned with ethanol and fresh water using papertowels or kimwipes. All of this equipment was contained in one plastic snap top bin after the experiment and labeled with “C13 use only” and kept in storage in the lab. This equipment will not be used for any use that contaminates other surfaces or seawater.
- All seawater used in the experiment was kept after the experiment in plastic screw top Nalgene bottles labeled “C13 waste seawater” and kept in a snap top plastic bin in the lab. These will be transported back to the US at a later date for proper disposal. We cannot dispose of in Moorea because all waste water has the potential to drain back into the ocean.
- As an extra precaution, all equipment and surfaces were thoroughly cleaned with ethanol and wipes after the experiment.
- All stock C13 sodium bicarbonate was contained in triple layer plastic bags and a separate container in the lab. This will be transported back to URI.
Set up
We set up three bins as water baths each on a 15 stir plate (as used for P:R measurements). The height of the water was set up so that 120mL glass jars containing samples (described below), will be submerged partially to control temperature.
Each bin/water bath contains heaters controlled with an Apex system, a recirculuating pump, and AquaIllumination LED lights. The temperature and light was controlled as described above for P and R measurements.
Hobo TidBit loggers were launched in each water bin logging every 10 minutes, starting at 12:30 (SSN 21723477, 21723473, 21723472).



We optimized the light exposure to be even across all samples. We did this by independently controlling light intensity above each bin. We needed lower intensity in the middle bin and higher intensity in the edge bins, for example, to make an even light field. Mean light values were 511 in bin 1 (left), 517 in bin 2 (middle), and 511 in bin 3 (right). Bin 1 lights were at 90% (all channels, no UV or moonlight), bin 2 were at 70% and bin 3 lights were at 100%. Light measurements were collected at each of the 15 locations in each bin.
Bins were randomly assigned a temperature treatment, and these were randomly re arranged for each day of incubations. Today, Bin 1 is 27C, Bin 2 is 30C, and Bin 3 is 33C. Each bin will have 15 jars with n=2 adults per species and n=3 recruits per species. Samples were chosen by randomly selecting 2 adults per species and picking a small, medium, and large recruit of each species.
Target incubation time is between 5 and 6 hours to match incubation time for PR measurements and conduct short term incubations to prevent metabolism of C13 stored in lipids as in previous literature.
Solutions
Isotopes were made at a concentration of 4 mM enrichment of sodium bicarbonate following previous literature. For today, I made 5.474 L of solution to supply all samples needed (45 samples x 120 mL (plus a little extra). This was the recipe today:
5 x 1000 mL 0.2 µm FSW and 340 mg C13
1 x 474 mL 0.2 µm FSW and 161.16 mg C13
This was calculated using the desired molarity and the molar mass of the sodium bicarbonate. Here is the formula:
4 mM / 1000 x 5475 mL / 1000 x 85 g/mol = 1.8616 g total C13, which divides to 340 mg per L of FSW.
The measured pH of the solution was 8.0-8.2. The solution was not acidic and therefore there was no need for balacing with NaOH solution.
Run 1
Starting at 14:20, a randomly selected sample from each batch was added into a 120 mL glass jar on a plug stand with a stir bar (see photos above). Ariana then filled the jar with C13 FSW following protocols listed above. The jar was then placed in the water bath bin. Bins were loaded between 14:20-14:56.
After all samples were loaded, temperature was ramped to target over a 15-20 minute period and jars reached target temperature by approximately 30 minutes. Stirring was set at a rate of 380 as done for PR measurements. Corals were at 5 cm depth.
Temperature was recorded several times throughout the run to verify treatment temperature and will also be verified using logger data.
Sample location was randomly rearranged within bin to account for any light variation at 17:00-17:30.
Sample and run metadata are on GitHub here.
Sampling
We started sampling corals at 19:35 after 5 hours of incubation. We noticed that at 33°C, almost all Acropora samples, both adult and recruit samples, had cloudy water. No mortality or tissue loss seen. No cloudy water in other species. Perhaps this is a sign of stress?
To sample, fragments were removed with gloved hands by Ariana and placed in a whirl pack with the whirl pack opened and held by another person to avoid any C13 touching the outside of the whirl pack. The whirl pack was then closed, the time recorded, and submerged in liquid nitrogen in <30s from removal from treatment.
After all samples were collected (staggered time to have samples have equal incubation time), samples were removed from liquid nitrogen and stored in the BIOCODE -40°C.
Acropora larval care
Acropora larvae looked good today in the squaricals and the coolers. We did water changes and cleaned banjos and filters for the Acropora larvae. Given that we now have both Pocillopora and Acropora larvae, here is my plan for what I would like to do for larval measurements. This will give us larval, recruit, and adult lifestages in this experiment, woo hoo! This will also give us aposymbiotic species and symbiotic species comparisons!
-
P&R and RNA: Incubate in glass vials at 27, 30, and 33°C as done for adults and measure metabolic rates in the light (photosynthesis, we expect 0 here for ACR as they have no symbionts) and dark (respiration) after 5 hours of exposure. Additional vials will be sampled for RNA as done for the adults. Run n=10 pools of 5 larvae per species per temperature. We will need to conduct a PI curve for Pocillopora larvae before measurements to confirm saturating irradiance, which will likely be lower for larvae.
-
Trial symbiont infections: We will put ACR larvae in plates and vials to trial symbiont infections. If successful, we could run measurements both on aposymbiotic and symbiotic larvae. We don’t have a lot of time to trial this, so I’m not sure we will have enough for measurements.
-
Stable isotope metabolomics: Incubate POC larvae at 27, 30, and 33°C as done for the adults but in glass vials. Only incubate POC larvae as we do not expect ACR larvae to take up symbionts by that point.
I’ll work on planning the specifics of these measurements next!
Monday - November 13, 2023
Morning spawning
There was a little POC spawning this morning, but not as much as yesterday. For our experiments for this project we will use the 20231112 cohort when larval measurements take place.
Isotopes day 2 - run 2 and dark run
Run 2
We ran the second and final C13 samples for isotope metabolomics today. The protocol was followed as described yesterday. The samples were loaded from 10:18-11:01 with a 15-20 min temperature ramp for the water bath. Samples were rearranged within tank at 13:00.
Sample and run metadata are on GitHub here.
Corals were sampled from 15:30-16:45 (5-5.5 hr incubation) as described for yesterday.
Run 3 (dark controls)
In the evening, we ran dark controls for C13 metabolomics. The purpose of the dark controls is to verify that photosynthates are enriched in C13 only in the light, when photosynthesis is occuring.
To do this, we exposed n=1 coral per group (lifestage x species) to 27°C in the dark. All other aspects of the incubation were run as described for light samples. Samples were loaded at 16:30 and sampled at 21:30.
Larval care
Larvae were cleaned, stirred, and maintained. They are healthy and swimming!
Trial symbiont infections
I set up some trial symbiont infections by airbrushing 1 Acropora hyacinthus fragment and isolating the pellet in filtered seawater. Tissue slurry was spun at 4000 rmp for 4 minutes, rinsed in FSW, and spun again before resuspending in FSW. I added some larvae to 6 well plates with a drop of symbiont concentrate and left over night. I’ll check tomorrow to see if they took up any symbionts using the fluorescent scope in the lab. Larvae placed in incubator at 26°C overnight.
Tuesday - November 14, 2023
Morning spawning
More spawning today! See Pierrick and Hollie’s notebooks for spawning and rearing information.
Larval care
Larvae care continued as described for previous days.
Planning larval experiments
We will conduct larval experiments tomorrow. I labeled tubes and vials today in preparation. We will use the 20231112 cohort of POC larvae and larvae from the squaricals for Acropora hyacinthus. We will do staggered start times for larvae that will be incubated and measured for P&R first. We will start the vials for RNA and metabolomics later so that we have time to do all of the measurements in the same day.
Today we incubated some ACR larvae in 20 mL glass vials at 33°C and 27°C for 5 hours to make sure the larvae survived. They all looked happy at 5 hours and even survived overnight!
We will do the following replication:
- P&R: n=10 wells per species per temperature (1 plate per species); n=10 blanks per temperature. n=60 POC larvae per well (based on HP and PH initial measurements for thresholds of larvae needed to get a P signal) and n=6 ACR per well (based on last year’s data).
- RNA: n=6 vials per species per temperature (200 POC per vial, 100 ACR per vial). Sample POC into 200 µL DNA/RNA shield for POC and 500 µL for ACR to account for more biomass in ACR.
- Metabolomics: n=6 vials of POC per temperature + 3 vials POC in the dark (black glass jars) at 27°C only. 200 larvae per vial. We will pour the larvae through a cell strainer, rinse the larvae in FSW, concentrate in a cell strainer, and sample in a screw top tube into liquid nitrogen to sample.
- Physiology: n=3 tubes per species snap frozen. 100 ACR and 200 POC for these samples.
We will sample larvae by pouring vial of water with larvae through a cell strainer and concentrating with a glass pipette into a screw top 1.5 mL tube and sample either into RNA DNA shield or snap frozen in liquid nitrogen. All samples will be stored at -40°C. We will also collect n=3 tubes per species for larval size images and fix in 37% formalin and store at 4°C.
We will use a total of 11,000 POC larvae and ~5,500 ACR larvae.
Tubes are labeled as follows:
- M1-M21 (POC metabolomics)
- C1-C18 (physiology)
- E1-E6 (extra tubes for trialing extractions)
- A1-A18 (Acropora RNA)
- P1-P18 (Pocillopora RNA)
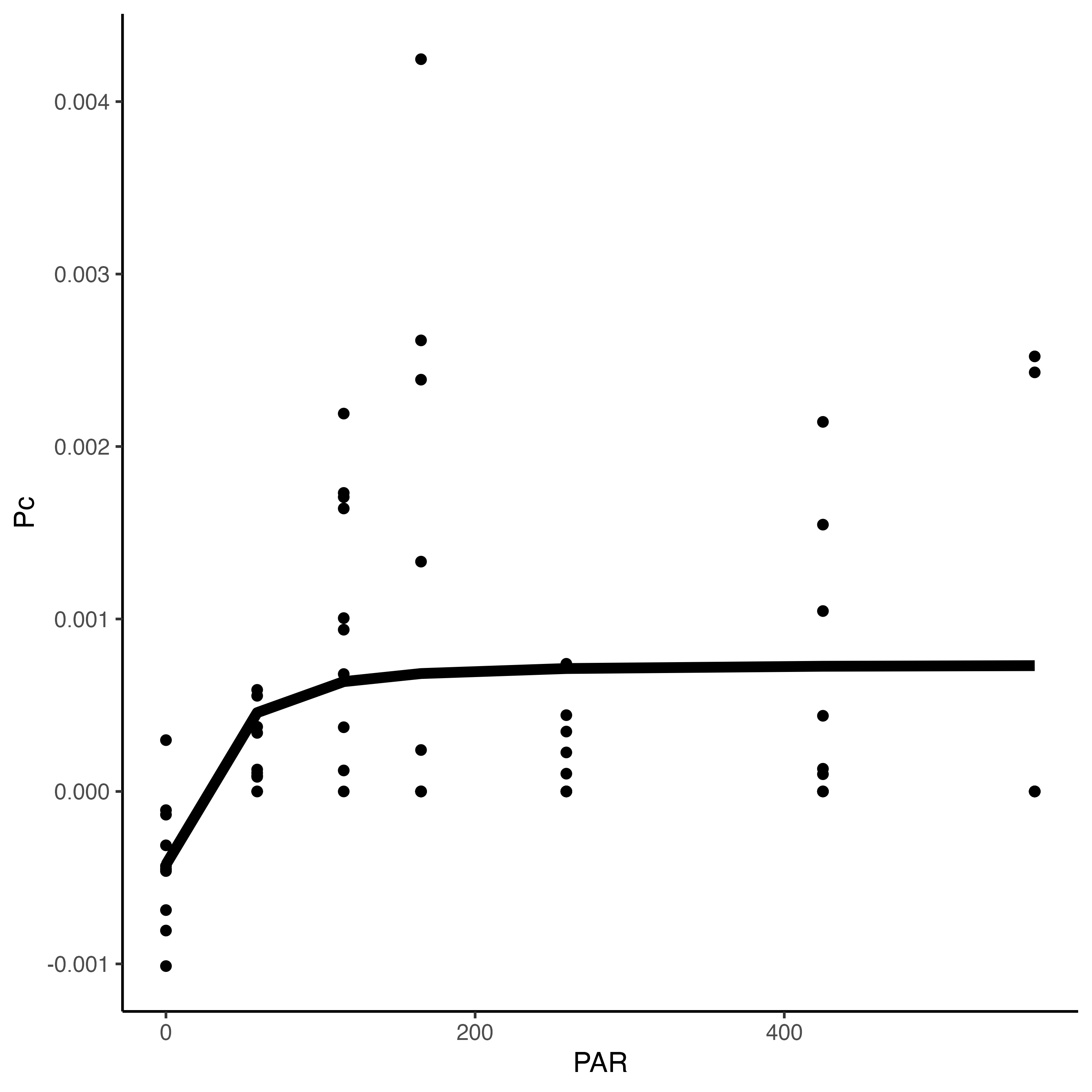
PI curves
Hollie and Pierrick ran a PI curve with n=60 POC larvae per well and we found saturating irradiance to be ~50 PAR. We will run incubations at 150 PAR to meet saturating irradiance.
Data files for the PI curves can be found here. Here is the preliminary PI curve and parameter estimates.

Daily measurements
Daily measurements were collected at 16:30.
Wednesday - November 15, 2023
POC and ACR larval sampling: respirometry, RNA, metabolomics, physiology, and size
Today we ran our larval experiments! All larval work will occur today so that there are not developmental effects. POC larvae are 3-4 days post fertilization. These larvae develop into swimming planulae within about 6-9 hours post fertilization! They develop very quickly. All notes on larval rearing and development are in Hollie’s and Pierrick’s notebooks (NSF RAPID project).
Loading vials at density
To load larvae into vials for incubations and measurements, we collected a concentrated stock of larvae of each species by scooping into squarical containers (n=12 containers for ACR and n=1 container for POC) with mesh scoops (35 µm for POC and 253 µm for ACR) and concentrating with a transfer pipette into 50 mL falcon tubes (3-4 per species). We then took 6 subsampled counts in a known volume (0.5-1 mL) and counted the number of larvae in each subsample. This was then used to calculate the density of the full stock falcon tube.
The desired volume to obtain the desired number of larvae was calculated and this volume was added into each vial.
Vials twere then filled with 0.2 µm filtered seawater with the exception of stable isotope metabolomics incubation vials, which were filled with 4mM C13 sodium bicarbonate solution as described above for the adult/recruit experiments.
Here are the samples we ran today and the full metadata:
| tube_id | species | temp | type | number_larvae | preservation | storage_location | freezer |
|---|---|---|---|---|---|---|---|
| M1 | Pocillopora | 27 | C13 | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| M2 | Pocillopora | 27 | C13 | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| M3 | Pocillopora | 27 | C13 | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| M4 | Pocillopora | 27 | C13 | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| M5 | Pocillopora | 27 | C13 | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| M6 | Pocillopora | 27 | C13 | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| M7 | Pocillopora | 30 | C13 | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| M8 | Pocillopora | 30 | C13 | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| M9 | Pocillopora | 30 | C13 | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| M10 | Pocillopora | 30 | C13 | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| M11 | Pocillopora | 30 | C13 | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| M12 | Pocillopora | 30 | C13 | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| M13 | Pocillopora | 33 | C13 | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| M14 | Pocillopora | 33 | C13 | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| M15 | Pocillopora | 33 | C13 | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| M16 | Pocillopora | 33 | C13 | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| M17 | Pocillopora | 33 | C13 | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| M18 | Pocillopora | 33 | C13 | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| M19 | Pocillopora | 27 | C13 Dark | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| M20 | Pocillopora | 27 | C13 Dark | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| M21 | Pocillopora | 27 | C13 Dark | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| P1 | Pocillopora | 27 | RNA | 200 | 250 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| P2 | Pocillopora | 27 | RNA | 200 | 250 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| P3 | Pocillopora | 27 | RNA | 200 | 250 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| P4 | Pocillopora | 27 | RNA | 200 | 250 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| P5 | Pocillopora | 27 | RNA | 200 | 250 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| P6 | Pocillopora | 27 | RNA | 200 | 250 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| P7 | Pocillopora | 30 | RNA | 200 | 250 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| P8 | Pocillopora | 30 | RNA | 200 | 250 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| P9 | Pocillopora | 30 | RNA | 200 | 250 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| P10 | Pocillopora | 30 | RNA | 200 | 250 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| P11 | Pocillopora | 30 | RNA | 200 | 250 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| P12 | Pocillopora | 30 | RNA | 200 | 250 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| P13 | Pocillopora | 33 | RNA | 200 | 250 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| P14 | Pocillopora | 33 | RNA | 200 | 250 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| P15 | Pocillopora | 33 | RNA | 200 | 250 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| P16 | Pocillopora | 33 | RNA | 200 | 250 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| P17 | Pocillopora | 33 | RNA | 200 | 250 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| P18 | Pocillopora | 33 | RNA | 200 | 250 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| A1 | Acropora | 27 | RNA | 100 | 500 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| A2 | Acropora | 27 | RNA | 100 | 500 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| A3 | Acropora | 27 | RNA | 100 | 500 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| A4 | Acropora | 27 | RNA | 100 | 500 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| A5 | Acropora | 27 | RNA | 100 | 500 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| A6 | Acropora | 27 | RNA | 100 | 500 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| A7 | Acropora | 30 | RNA | 100 | 500 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| A8 | Acropora | 30 | RNA | 100 | 500 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| A9 | Acropora | 30 | RNA | 100 | 500 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| A10 | Acropora | 30 | RNA | 100 | 500 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| A11 | Acropora | 30 | RNA | 100 | 500 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| A12 | Acropora | 30 | RNA | 100 | 500 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| A13 | Acropora | 33 | RNA | 100 | 500 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| A14 | Acropora | 33 | RNA | 100 | 500 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| A15 | Acropora | 33 | RNA | 100 | 500 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| A16 | Acropora | 33 | RNA | 100 | 500 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| A17 | Acropora | 33 | RNA | 100 | 500 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| A18 | Acropora | 33 | RNA | 100 | 500 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| C1 | Acropora | 27 | Physiology | 100 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| C2 | Acropora | 27 | Physiology | 100 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| C3 | Acropora | 27 | Physiology | 100 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| C4 | Acropora | 30 | Physiology | 100 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| C5 | Acropora | 30 | Physiology | 100 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| C6 | Acropora | 30 | Physiology | 100 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| C7 | Acropora | 33 | Physiology | 100 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| C8 | Acropora | 33 | Physiology | 100 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| C9 | Acropora | 33 | Physiology | 100 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| C10 | Pocillopora | 27 | Physiology | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| C11 | Pocillopora | 27 | Physiology | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| C12 | Pocillopora | 27 | Physiology | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| C13 | Pocillopora | 30 | Physiology | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| C14 | Pocillopora | 30 | Physiology | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| C15 | Pocillopora | 30 | Physiology | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| C16 | Pocillopora | 33 | Physiology | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| C17 | Pocillopora | 33 | Physiology | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| C18 | Pocillopora | 33 | Physiology | 200 | Snap frozen | Box 6 | Molecular -40C freezer, shelf 4 |
| E1 | Acropora | 27 | Extra | 100 | 500 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| E2 | Acropora | 27 | Extra | 100 | 500 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| E3 | Acropora | 27 | Extra | 100 | 500 uL shield | Box 4 | Molecular -40C freezer, shelf 4 |
| E4 | Pocillopora | 27 | Extra | 200 | 250 uL shield | Box 6 | Molecular -40C freezer, shelf 4 |
| E5 | Pocillopora | 27 | Extra | 200 | 250 uL shield | Box 6 | Molecular -40C freezer, shelf 4 |
| E6 | Pocillopora | 27 | Extra | 200 | 250 uL shield | Box 6 | Molecular -40C freezer, shelf 4 |
This metadata file is available on GitHub here.
Incubations
Larvae were added using density calculations described above to 20 mL glass scintillation vials with white caps partially (~90%) submerged in water baths. Dark isotope samples were loaded in 120 mL black glass jars. Vials were loaded from 13:45-16:00.
Larvae that were incubated for P&R measurements were loaded into 3 replicate falcon tubes per temperature. Larvae will be pulled from these falcon tubes for SDR measurements. These vials were loaded first from 08:00-10:00.
Temperatures are recorded using loggers as done for the isotope measurements and manually measured using a thermometer throughout the incubations. Logger 21723473 was in Bin 1 at 30°C; Logger 21723472 was in Bin 2 at 33°C; Logger 21723477 was in Bin 3 at 27°C.
To achieve an even light field of 150 PAR as indicated by our PI curves, lights were set at 25% for Bin 1, 20% for Bin 2, and 25% for Bin 3 to achieve 150-160 PAR in the center of each bin. Once throughout the vials were rearranged within each bin.
Notebook images with manual temperature measurements for all experiments are on Google Drive.
Here is an image of our larval incubation set up from today.

Respirometry
The first SDR run was conducted from 15:57-16:35. This was later than originally planned, but was the earliest we could conduct the measurements. We conducted a 100% air saturated seawater calibration before this run using 0.2µm seawater that was bubbled for 30 minutes before calibration. Sensors were reading a bit high before the calibration (~120%) and were reading 100% afterwards.
SDR Run 1 was run at 27°C with larvae incubated at 30°C. We will use 6 larvae per well for ACR and 60 larvae per well for POC. POC larvae are counted under a fluorescent scope/blue light into small pools of water that are then added into the wells of the glass plate.
Subsequently, we conducted Run #2 at 27°C (18:05-18:42) and Run #3 at 33°C (21:08-21:50).
SDR runs followed protocols established in the Putnam Lab and using protocols developed for the Hawaii 2023 project. We ran measurements using a pair of SDR readers (PreSens) and glass plates with oxygen dot sensors (80 µL; Loligo).
Briefly, we followed the following proceedure for each run:
- n=10 larvae were loaded into a plate with n=10 FSW blanks. We loaded ACR in one plate and POC in another. Larvae were loaded and the well was filled with 0.2 µm FSW and sealed with glass cover slips.
- Plates were placed on SDR readers in benchtop incubators at the treatment temperature. We first ran a light exposure period for 15-20 minutes at 18% AI 16HD LED light intensity (150 PAR) for photosynthesis.
- We then turned the lights off to measure respiration for 12-15 minutes (light enhanced respiration).
Note that becuase ACR does not have symbionts, we expect that there will be net oxygen depletion in ACR during the light period. We will extract oxygen evolution rates for ACR respiration from the first light period. This is because the well will reach oxygen depletion by the time that we enter the dark period, which will result in lower than actual respiration rates.
Here is the run and sample metadata for SDR runs #1-3:
| Date | Run | Plate | SDR | Temperature | Light_Level | Light_Value | TimeStart | TimeStop | TimeInterval | IntervalStart | IntervalStop | Notes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 20231115 | 1 | 1 | 641 | 30 | 1 | 150 | 15:57 | 16:18 | 0:21 | 0 | 21 | |
| 20231115 | 1 | 1 | 641 | 30 | 2 | 0 | 16:20 | 16:35 | 0:15 | 23 | 38 | |
| 20231115 | 1 | 2 | 710 | 30 | 1 | 150 | 15:57 | 16:18 | 0:21 | 0 | 21 | |
| 20231115 | 1 | 2 | 710 | 30 | 2 | 0 | 16:20 | 16:35 | 0:15 | 23 | 38 | |
| 20231115 | 2 | 3 | 641 | 27 | 1 | 150 | 18:05 | 18:25 | 0:20 | 0 | 20 | |
| 20231115 | 2 | 3 | 641 | 27 | 2 | 0 | 18:27 | 18:42 | 0:15 | 22 | 37 | |
| 20231115 | 2 | 4 | 710 | 27 | 1 | 150 | 18:05 | 18:25 | 0:20 | 0 | 20 | |
| 20231115 | 2 | 4 | 710 | 27 | 2 | 0 | 18:27 | 18:42 | 0:15 | 22 | 37 | |
| 20231115 | 3 | 5 | 641 | 33 | 1 | 150 | 21:08 | 21:28 | 0:20 | 0 | 20 | |
| 20231115 | 3 | 5 | 641 | 33 | 2 | 0 | 21:30 | 21:50 | 0:20 | 22 | 42 | |
| 20231115 | 3 | 6 | 710 | 33 | 1 | 150 | 21:08 | 21:28 | 0:20 | 0 | 20 | |
| 20231115 | 3 | 6 | 710 | 33 | 2 | 0 | 21:30 | 21:50 | 0:20 | 22 | 42 |
| Date | Sample.ID | Chamber.ID | SDR | Temperature | Species | Type | Salinity | Plate | Run | Volume | Org.Number | Notes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 20231115 | 1 | A1 | 641 | 30 | Pocillopora | Sample | 36 | 1 | 1 | 0.00008 | 60 | |
| 20231115 | 7 | B1 | 641 | 30 | Pocillopora | Sample | 36 | 1 | 1 | 0.00008 | 60 | |
| 20231115 | 13 | C1 | 641 | 30 | NA | Blank | 36 | 1 | 1 | 0.00008 | NA | |
| 20231115 | 19 | D1 | 641 | 30 | NA | Blank | 36 | 1 | 1 | 0.00008 | NA | |
| 20231115 | 2 | A2 | 641 | 30 | NA | Blank | 36 | 1 | 1 | 0.00008 | NA | |
| 20231115 | 8 | B2 | 641 | 30 | NA | Blank | 36 | 1 | 1 | 0.00008 | NA | |
| 20231115 | 14 | C2 | 641 | 30 | NA | Blank | 36 | 1 | 1 | 0.00008 | NA | |
| 20231115 | 20 | D2 | 641 | 30 | Pocillopora | Sample | 36 | 1 | 1 | 0.00008 | 60 | |
| 20231115 | 3 | A3 | 641 | 30 | NA | Blank | 36 | 1 | 1 | 0.00008 | NA | |
| 20231115 | 9 | B3 | 641 | 30 | Pocillopora | Sample | 36 | 1 | 1 | 0.00008 | 60 | |
| 20231115 | 15 | C3 | 641 | 30 | Pocillopora | Sample | 36 | 1 | 1 | 0.00008 | 60 | |
| 20231115 | 21 | D3 | 641 | 30 | NA | Blank | 36 | 1 | 1 | 0.00008 | NA | |
| 20231115 | 4 | A4 | 641 | 30 | NA | Blank | 36 | 1 | 1 | 0.00008 | NA | |
| 20231115 | 10 | B4 | 641 | 30 | Pocillopora | Sample | 36 | 1 | 1 | 0.00008 | 60 | |
| 20231115 | 16 | C4 | 641 | 30 | Pocillopora | Sample | 36 | 1 | 1 | 0.00008 | 60 | |
| 20231115 | 22 | D4 | 641 | 30 | NA | Blank | 36 | 1 | 1 | 0.00008 | NA | |
| 20231115 | 5 | A5 | 641 | 30 | Pocillopora | Sample | 36 | 1 | 1 | 0.00008 | 60 | |
| 20231115 | 11 | B5 | 641 | 30 | NA | Blank | 36 | 1 | 1 | 0.00008 | NA | |
| 20231115 | 17 | C5 | 641 | 30 | NA | Blank | 36 | 1 | 1 | 0.00008 | NA | |
| 20231115 | 23 | D5 | 641 | 30 | NA | Blank | 36 | 1 | 1 | 0.00008 | NA | |
| 20231115 | 6 | A6 | 641 | 30 | NA | Blank | 36 | 1 | 1 | 0.00008 | NA | |
| 20231115 | 12 | B6 | 641 | 30 | Pocillopora | Sample | 36 | 1 | 1 | 0.00008 | 60 | |
| 20231115 | 18 | C6 | 641 | 30 | Pocillopora | Sample | 36 | 1 | 1 | 0.00008 | 60 | |
| 20231115 | 24 | D6 | 641 | 30 | NA | Blank | 36 | 1 | 1 | 0.00008 | NA | |
| 20231115 | 1 | A1 | 740 | 30 | Acropora | Sample | 36 | 2 | 1 | 0.00008 | 6 | |
| 20231115 | 7 | B1 | 740 | 30 | NA | Blank | 36 | 2 | 1 | 0.00008 | NA | |
| 20231115 | 13 | C1 | 740 | 30 | Acropora | Sample | 36 | 2 | 1 | 0.00008 | 6 | |
| 20231115 | 19 | D1 | 740 | 30 | NA | Blank | 36 | 2 | 1 | 0.00008 | NA | |
| 20231115 | 2 | A2 | 740 | 30 | NA | Blank | 36 | 2 | 1 | 0.00008 | NA | |
| 20231115 | 8 | B2 | 740 | 30 | Acropora | Sample | 36 | 2 | 1 | 0.00008 | 6 | |
| 20231115 | 14 | C2 | 740 | 30 | NA | Blank | 36 | 2 | 1 | 0.00008 | NA | |
| 20231115 | 20 | D2 | 740 | 30 | NA | Blank | 36 | 2 | 1 | 0.00008 | NA | |
| 20231115 | 3 | A3 | 740 | 30 | NA | Blank | 36 | 2 | 1 | 0.00008 | NA | |
| 20231115 | 9 | B3 | 740 | 30 | Acropora | Sample | 36 | 2 | 1 | 0.00008 | 6 | |
| 20231115 | 15 | C3 | 740 | 30 | NA | Blank | 36 | 2 | 1 | 0.00008 | NA | |
| 20231115 | 21 | D3 | 740 | 30 | Acropora | Sample | 36 | 2 | 1 | 0.00008 | 6 | |
| 20231115 | 4 | A4 | 740 | 30 | Acropora | Sample | 36 | 2 | 1 | 0.00008 | 6 | |
| 20231115 | 10 | B4 | 740 | 30 | NA | Blank | 36 | 2 | 1 | 0.00008 | NA | |
| 20231115 | 16 | C4 | 740 | 30 | NA | Blank | 36 | 2 | 1 | 0.00008 | NA | |
| 20231115 | 22 | D4 | 740 | 30 | NA | Blank | 36 | 2 | 1 | 0.00008 | NA | |
| 20231115 | 5 | A5 | 740 | 30 | NA | Blank | 36 | 2 | 1 | 0.00008 | NA | |
| 20231115 | 11 | B5 | 740 | 30 | NA | Blank | 36 | 2 | 1 | 0.00008 | NA | |
| 20231115 | 17 | C5 | 740 | 30 | Acropora | Sample | 36 | 2 | 1 | 0.00008 | 6 | |
| 20231115 | 23 | D5 | 740 | 30 | Acropora | Sample | 36 | 2 | 1 | 0.00008 | 6 | |
| 20231115 | 6 | A6 | 740 | 30 | Acropora | Sample | 36 | 2 | 1 | 0.00008 | 6 | |
| 20231115 | 12 | B6 | 740 | 30 | Acropora | Sample | 36 | 2 | 1 | 0.00008 | 6 | |
| 20231115 | 18 | C6 | 740 | 30 | NA | Blank | 36 | 2 | 1 | 0.00008 | NA | |
| 20231115 | 24 | D6 | 740 | 30 | NA | Blank | 36 | 2 | 1 | 0.00008 | NA | |
| 20231115 | 1 | A1 | 641 | 27 | Acropora | Sample | 36 | 3 | 2 | 0.00008 | 6 | |
| 20231115 | 7 | B1 | 641 | 27 | Acropora | Sample | 36 | 3 | 2 | 0.00008 | 6 | |
| 20231115 | 13 | C1 | 641 | 27 | Acropora | Sample | 36 | 3 | 2 | 0.00008 | 6 | |
| 20231115 | 19 | D1 | 641 | 27 | NA | Blank | 36 | 3 | 2 | 0.00008 | NA | |
| 20231115 | 2 | A2 | 641 | 27 | NA | Blank | 36 | 3 | 2 | 0.00008 | NA | |
| 20231115 | 8 | B2 | 641 | 27 | Acropora | Sample | 36 | 3 | 2 | 0.00008 | 6 | |
| 20231115 | 14 | C2 | 641 | 27 | NA | Blank | 36 | 3 | 2 | 0.00008 | NA | |
| 20231115 | 20 | D2 | 641 | 27 | NA | Blank | 36 | 3 | 2 | 0.00008 | NA | |
| 20231115 | 3 | A3 | 641 | 27 | NA | Blank | 36 | 3 | 2 | 0.00008 | NA | |
| 20231115 | 9 | B3 | 641 | 27 | NA | Blank | 36 | 3 | 2 | 0.00008 | NA | |
| 20231115 | 15 | C3 | 641 | 27 | NA | Blank | 36 | 3 | 2 | 0.00008 | NA | |
| 20231115 | 21 | D3 | 641 | 27 | Acropora | Sample | 36 | 3 | 2 | 0.00008 | 6 | |
| 20231115 | 4 | A4 | 641 | 27 | Acropora | Sample | 36 | 3 | 2 | 0.00008 | 6 | |
| 20231115 | 10 | B4 | 641 | 27 | Acropora | Sample | 36 | 3 | 2 | 0.00008 | 6 | |
| 20231115 | 16 | C4 | 641 | 27 | NA | Blank | 36 | 3 | 2 | 0.00008 | NA | |
| 20231115 | 22 | D4 | 641 | 27 | Acropora | Sample | 36 | 3 | 2 | 0.00008 | 6 | |
| 20231115 | 5 | A5 | 641 | 27 | NA | Blank | 36 | 3 | 2 | 0.00008 | NA | |
| 20231115 | 11 | B5 | 641 | 27 | NA | Blank | 36 | 3 | 2 | 0.00008 | NA | |
| 20231115 | 17 | C5 | 641 | 27 | Acropora | Sample | 36 | 3 | 2 | 0.00008 | 6 | |
| 20231115 | 23 | D5 | 641 | 27 | NA | Blank | 36 | 3 | 2 | 0.00008 | NA | |
| 20231115 | 6 | A6 | 641 | 27 | NA | Blank | 36 | 3 | 2 | 0.00008 | NA | |
| 20231115 | 12 | B6 | 641 | 27 | Acropora | Sample | 36 | 3 | 2 | 0.00008 | 6 | |
| 20231115 | 18 | C6 | 641 | 27 | NA | Blank | 36 | 3 | 2 | 0.00008 | NA | |
| 20231115 | 24 | D6 | 641 | 27 | NA | Blank | 36 | 3 | 2 | 0.00008 | NA | |
| 20231115 | 1 | A1 | 740 | 27 | NA | Blank | 36 | 4 | 2 | 0.00008 | NA | |
| 20231115 | 7 | B1 | 740 | 27 | NA | Blank | 36 | 4 | 2 | 0.00008 | NA | |
| 20231115 | 13 | C1 | 740 | 27 | Pocillopora | Sample | 36 | 4 | 2 | 0.00008 | 60 | |
| 20231115 | 19 | D1 | 740 | 27 | NA | Blank | 36 | 4 | 2 | 0.00008 | NA | |
| 20231115 | 2 | A2 | 740 | 27 | Pocillopora | Sample | 36 | 4 | 2 | 0.00008 | 60 | |
| 20231115 | 8 | B2 | 740 | 27 | NA | Blank | 36 | 4 | 2 | 0.00008 | NA | |
| 20231115 | 14 | C2 | 740 | 27 | Pocillopora | Sample | 36 | 4 | 2 | 0.00008 | 60 | |
| 20231115 | 20 | D2 | 740 | 27 | NA | Blank | 36 | 4 | 2 | 0.00008 | NA | |
| 20231115 | 3 | A3 | 740 | 27 | NA | Blank | 36 | 4 | 2 | 0.00008 | NA | |
| 20231115 | 9 | B3 | 740 | 27 | Pocillopora | Sample | 36 | 4 | 2 | 0.00008 | 60 | |
| 20231115 | 15 | C3 | 740 | 27 | NA | Blank | 36 | 4 | 2 | 0.00008 | NA | |
| 20231115 | 21 | D3 | 740 | 27 | Pocillopora | Sample | 36 | 4 | 2 | 0.00008 | 60 | |
| 20231115 | 4 | A4 | 740 | 27 | Pocillopora | Sample | 36 | 4 | 2 | 0.00008 | 60 | |
| 20231115 | 10 | B4 | 740 | 27 | NA | Blank | 36 | 4 | 2 | 0.00008 | NA | |
| 20231115 | 16 | C4 | 740 | 27 | Pocillopora | Sample | 36 | 4 | 2 | 0.00008 | 60 | |
| 20231115 | 22 | D4 | 740 | 27 | NA | Blank | 36 | 4 | 2 | 0.00008 | NA | |
| 20231115 | 5 | A5 | 740 | 27 | Pocillopora | Sample | 36 | 4 | 2 | 0.00008 | 60 | |
| 20231115 | 11 | B5 | 740 | 27 | NA | Blank | 36 | 4 | 2 | 0.00008 | NA | |
| 20231115 | 17 | C5 | 740 | 27 | NA | Blank | 36 | 4 | 2 | 0.00008 | NA | |
| 20231115 | 23 | D5 | 740 | 27 | NA | Blank | 36 | 4 | 2 | 0.00008 | NA | |
| 20231115 | 6 | A6 | 740 | 27 | NA | Blank | 36 | 4 | 2 | 0.00008 | NA | |
| 20231115 | 12 | B6 | 740 | 27 | Pocillopora | Sample | 36 | 4 | 2 | 0.00008 | 60 | |
| 20231115 | 18 | C6 | 740 | 27 | NA | Blank | 36 | 4 | 2 | 0.00008 | NA | |
| 20231115 | 24 | D6 | 740 | 27 | Pocillopora | Sample | 36 | 4 | 2 | 0.00008 | 60 | |
| 20231115 | 1 | A1 | 641 | 33 | NA | Blank | 36 | 5 | 3 | 0.00008 | NA | |
| 20231115 | 7 | B1 | 641 | 33 | Acropora | Sample | 36 | 5 | 3 | 0.00008 | 6 | |
| 20231115 | 13 | C1 | 641 | 33 | NA | Blank | 36 | 5 | 3 | 0.00008 | NA | |
| 20231115 | 19 | D1 | 641 | 33 | Acropora | Sample | 36 | 5 | 3 | 0.00008 | 6 | |
| 20231115 | 2 | A2 | 641 | 33 | Acropora | Sample | 36 | 5 | 3 | 0.00008 | 6 | |
| 20231115 | 8 | B2 | 641 | 33 | NA | Blank | 36 | 5 | 3 | 0.00008 | NA | |
| 20231115 | 14 | C2 | 641 | 33 | Acropora | Sample | 36 | 5 | 3 | 0.00008 | 6 | |
| 20231115 | 20 | D2 | 641 | 33 | Acropora | Sample | 36 | 5 | 3 | 0.00008 | 6 | |
| 20231115 | 3 | A3 | 641 | 33 | Acropora | Sample | 36 | 5 | 3 | 0.00008 | 6 | |
| 20231115 | 9 | B3 | 641 | 33 | NA | Blank | 36 | 5 | 3 | 0.00008 | NA | |
| 20231115 | 15 | C3 | 641 | 33 | NA | Blank | 36 | 5 | 3 | 0.00008 | NA | |
| 20231115 | 21 | D3 | 641 | 33 | NA | Blank | 36 | 5 | 3 | 0.00008 | NA | |
| 20231115 | 4 | A4 | 641 | 33 | NA | Blank | 36 | 5 | 3 | 0.00008 | NA | |
| 20231115 | 10 | B4 | 641 | 33 | Acropora | Sample | 36 | 5 | 3 | 0.00008 | 6 | |
| 20231115 | 16 | C4 | 641 | 33 | NA | Blank | 36 | 5 | 3 | 0.00008 | NA | |
| 20231115 | 22 | D4 | 641 | 33 | Acropora | Sample | 36 | 5 | 3 | 0.00008 | 6 | |
| 20231115 | 5 | A5 | 641 | 33 | NA | Blank | 36 | 5 | 3 | 0.00008 | NA | |
| 20231115 | 11 | B5 | 641 | 33 | NA | Blank | 36 | 5 | 3 | 0.00008 | NA | |
| 20231115 | 17 | C5 | 641 | 33 | NA | Blank | 36 | 5 | 3 | 0.00008 | NA | |
| 20231115 | 23 | D5 | 641 | 33 | NA | Blank | 36 | 5 | 3 | 0.00008 | NA | |
| 20231115 | 6 | A6 | 641 | 33 | NA | Blank | 36 | 5 | 3 | 0.00008 | NA | |
| 20231115 | 12 | B6 | 641 | 33 | Acropora | Sample | 36 | 5 | 3 | 0.00008 | 6 | |
| 20231115 | 18 | C6 | 641 | 33 | NA | Blank | 36 | 5 | 3 | 0.00008 | NA | |
| 20231115 | 24 | D6 | 641 | 33 | Acropora | Sample | 36 | 5 | 3 | 0.00008 | 6 | |
| 20231115 | 1 | A1 | 740 | 33 | Pocillopora | Sample | 36 | 6 | 3 | 0.00008 | 60 | |
| 20231115 | 7 | B1 | 740 | 33 | NA | Blank | 36 | 6 | 3 | 0.00008 | NA | |
| 20231115 | 13 | C1 | 740 | 33 | Pocillopora | Sample | 36 | 6 | 3 | 0.00008 | 60 | |
| 20231115 | 19 | D1 | 740 | 33 | NA | Blank | 36 | 6 | 3 | 0.00008 | NA | |
| 20231115 | 2 | A2 | 740 | 33 | NA | Blank | 36 | 6 | 3 | 0.00008 | NA | |
| 20231115 | 8 | B2 | 740 | 33 | Pocillopora | Sample | 36 | 6 | 3 | 0.00008 | 60 | |
| 20231115 | 14 | C2 | 740 | 33 | NA | Blank | 36 | 6 | 3 | 0.00008 | NA | |
| 20231115 | 20 | D2 | 740 | 33 | Pocillopora | Sample | 36 | 6 | 3 | 0.00008 | 60 | |
| 20231115 | 3 | A3 | 740 | 33 | Pocillopora | Sample | 36 | 6 | 3 | 0.00008 | 60 | |
| 20231115 | 9 | B3 | 740 | 33 | Pocillopora | Sample | 36 | 6 | 3 | 0.00008 | 60 | |
| 20231115 | 15 | C3 | 740 | 33 | NA | Blank | 36 | 6 | 3 | 0.00008 | NA | |
| 20231115 | 21 | D3 | 740 | 33 | Pocillopora | Sample | 36 | 6 | 3 | 0.00008 | 60 | |
| 20231115 | 4 | A4 | 740 | 33 | NA | Blank | 36 | 6 | 3 | 0.00008 | NA | |
| 20231115 | 10 | B4 | 740 | 33 | NA | Blank | 36 | 6 | 3 | 0.00008 | NA | |
| 20231115 | 16 | C4 | 740 | 33 | Pocillopora | Sample | 36 | 6 | 3 | 0.00008 | 60 | |
| 20231115 | 22 | D4 | 740 | 33 | NA | Blank | 36 | 6 | 3 | 0.00008 | NA | |
| 20231115 | 5 | A5 | 740 | 33 | NA | Blank | 36 | 6 | 3 | 0.00008 | NA | |
| 20231115 | 11 | B5 | 740 | 33 | Pocillopora | Sample | 36 | 6 | 3 | 0.00008 | 60 | |
| 20231115 | 17 | C5 | 740 | 33 | NA | Blank | 36 | 6 | 3 | 0.00008 | NA | |
| 20231115 | 23 | D5 | 740 | 33 | NA | Blank | 36 | 6 | 3 | 0.00008 | NA | |
| 20231115 | 6 | A6 | 740 | 33 | NA | Blank | 36 | 6 | 3 | 0.00008 | NA | |
| 20231115 | 12 | B6 | 740 | 33 | NA | Blank | 36 | 6 | 3 | 0.00008 | NA | |
| 20231115 | 18 | C6 | 740 | 33 | Pocillopora | Sample | 36 | 6 | 3 | 0.00008 | 60 | |
| 20231115 | 24 | D6 | 740 | 33 | NA | Blank | 36 | 6 | 3 | 0.00008 | NA |
All data files can be found on GitHub here.
RNA, metabolomics, physiology
Larvae were loaded into vials for temperature treatment incubations and were sampled for RNA, stable isotope metabolomics, and physiology, as done for the adult/recruit experiments. We loaded larvae into vials (see replication and number of larvae above in the metadata table) as described above.
Metabolomics samples were loaded in solutions as described for adult/recruit samples. Metabolomics dark samples were dark adapted starting at 11:00am prior to incubations to prevent labeling of residual photosynthetic activity.
Vials were loaded between 13:30-16:00.
To sample, larvae were poured through a cell strainer held in 6-well plate wells with 0.2 µm FSW. Larvae were then gathered with as little water as possible using a glass pipette and transfered to respective labeled 1.5 mL tubes. Samples for RNA were preserved in RNA DNA shield (500 µL for ACR and 250 µL for POC). We used a different pipette and cell strainer for each treatment/species group.
Metabolomics samples were sampled in the same way using the isotope precautions and protocols described above. A dedicated set of pipettes, plates, and cell strainers were used for isotope samples. Larvae were snap frozen in liquid nitrogen.
Physiology samples were also sampled in this way and snap frozen. Sampling occured from 19:07 to 20:48 and staggered to obtain approx. 5 hour incubation times.
All samples were stored in the Molecular Lab -40°C freezer.
Larval size
We also collected tubes of 50-100 larvae of each species were collected after SDR runs and fixed in 37% formalin in Box 5 and stored at 4°C for later imaging.
Here are the samples we collected for larval size:
| tube_id | species | temp | type | number_larvae | preservation | storage_location | freezer |
|---|---|---|---|---|---|---|---|
| F1 | Acropora | 27 | Size | 50 | 200 uL 37% formalin | Box 5 | Molecular fridge 4C |
| F2 | Acropora | 27 | Size | 50 | 200 uL 37% formalin | Box 5 | Molecular fridge 4C |
| F3 | Pocillopora | 27 | Size | 50 | 200 uL 37% formalin | Box 5 | Molecular fridge 4C |
| F4 | Pocillopora | 27 | Size | 50 | 200 uL 37% formalin | Box 5 | Molecular fridge 4C |
| Poc 27 fixed | Pocillopora | 27 | Size | 50 | 200 uL 37% formalin | Box 5 | Molecular fridge 4C |
| Poc 30 fixed | Pocillopora | 30 | Size | 50 | 200 uL 37% formalin | Box 5 | Molecular fridge 4C |
| Poc 33 fixed | Pocillopora | 33 | Size | 50 | 200 uL 37% formalin | Box 5 | Molecular fridge 4C |
Box 4 contains A, P, and E tubes; Box 5 contains fixed F tubes; Box 6 contains M, C tubes.
Daily measurements
Daily measurements were collected at 08:30.
Thursday - November 16, 2023
Reva Atea
Ariana worked on the Reva Atea magazine today.
Larval rearing
Today Chloe cleaned water tables, took 10 extra tube samples of ACR into RNA DNA shield (Ex 1-10; 500 µL shield) and cleaned the adult/recruit tank. Chloe also cleaned all larval containers and set up some mesh bins for trial settlement! Larvae were added into mesh bottom bins with upside down aragonite plugs or pieces of coral rubble. We will monitor the larvae to see if we get settlement.
The temperature logger in the adult/recruit tank was moved at 15:16 to the new clean tank. We will use the remaining extra adult/recruit samples for a few more control/test runs.
Metabolomics control runs
We ran one metabolomics run today to run C12 and C13 controls and take some test samples of larvae for MALDI imaging. Here are the samples we ran today:
Adults: n=3 C12; n=3 C13 at 27°C
Larvae: n=3 C13 and n=3 FSW at 27°C and 33°C for POC; n=3 FSW at 27°C and 33°C for ACR
C12 controls were run to verify that the presence of C13 did not alter coral metabolism. To do this, we randomly selected n=1 adult per species for C12 exposure and C13 exposure. 4 mM solutions of C13 sodium bicarbonate (as described previouslyl 0.255 g + 750 mL FSW) and 4 mM solutions of C12 sodium bicarbonate (0.168 g + 500 mL FSW) were mixed and added into glass jars as previously describe. C12 samples were loaded into plastic respiration chambers that had no C13 exposure.
Light and temperature were controlled as described previously for adult/recruit experiments.
Samples were incubated from 15:00 and sampled as previously described at 20:30 (5.5 hours) into labeled whirl packs and snap frozen and stored at -40°C. Metadata can be found on GitHub here.
For larval samples, we loaded 20-30 ACR larvae into each ACR vial and 50-100 POC larvae. We used n=3 tubes with C13 label and n=3 in FSW at 27°C and 33°C for POC. We only ran FSW for ACR at 27°C and 33°C. This is because ACR do not have symbionts and therefore we are not interested in carbon tracing for the purpose of studying nutritional exchange.
These samples were sampled into 300 µL 4% CMC and snap frozen for potential use for future MALDI imaging with Jen Matthews.
Exposures were conducted as done before for larval incubuations at 150 PAR. Samples were loaded at 15:15 and sampled at 20:30 (about 5-5.5 hours).
Friday - November 17, 2023
RNA clipping of PR corals
Today we clipped biopsies for RNA from all corals run for P&R measurements on Nov 9-10, 2023. To do this, we removed a small batch of fragments from the freezer and kept them on ice. Fragments were then clipped for ~0.5 cm area (~10 polyp area). The biopsy was then put into a labeled 1.5 mL screw top tube in 500 µL RNA/DNA shield. The fragment was then returned to the freezer for physiology processing in a few days. Where fragments taken were larger, we added extra shield.
The samples were processed in a random order and gloves, tools, clippers, and bench space were sterilized with 70% ethanol, 10% bleach, and alcohol wipes between samples. Ariana and Lucy processed all samples between 13:50-17:00.
Samples were frozen in the -40°C freezer in Boxes 1 and 3.
Protocol followed that described on 11 November 2023.
The metadata from processing today is available on GitHub.
Sample inventory
Here are the boxes that we have in the freezer/fridge so far:
| Box | Preservative | Tubes | Transport |
|---|---|---|---|
| 1 | RNA/DNA Shield | DNA - PI Curves; DNA - PR | Room Temp |
| 2 | RNA/DNA Shield | DNA - Metabolomics corals | Room Temp |
| 3 | RNA/DNA Shield | DNA - PR Corals; DNA - Metabolomics corals | Room Temp |
| 4 | RNA/DNA Shield | POC & ACR Larvae RNA/DNA samples | Room Temp |
| 5 | Formalin | Larval size samples | Process in Moorea or Room Temp |
| 6 | Snap Frozen | Larval metabolomics, larval physiology, larval CMC samples | Dry shipper |
| Mesh bags x 2 | Snap Frozen | Isotope metabolomics fragments | Dry shipper |
Saturday - November 18, 2023
Airbrushing
We will start airbrushing corals from PR samples tomorrow. We will follow the established E5 protocol with the following modifications:
- Take a small batch of fragments out of the freezer and put in a cooler on ice
- Airbrush using an artists’ airbrush with cold 1X PBS buffer
- Airbrush by first using only air to get as much tissue as possible before adding PBS to concentrate the tissue. Airbrush into a plastic sandwich size bag or a whirl pack for very small fragments.
- Once the skeleton is clean, pour tissue slurry into a 5 mL or 15 mL, depending on the total amount of volume. We aimed for 3-15 mL total volume. Recruits had lower volume than adults for small recruits. We had 2 people airbrushing at a time.
- After airbrushing, slurry was homogenized using a Pro Scientific Tissue Shearer for ~15 sec. Homogenizer was cleaned with 70% ethanol and DI water between samples.
- From homogenized slurry, take 1 mL sample into a tube labeled with the coral ID + “S”. This is the symbiont tube.
- Put remaining homogenate into the freezer for later use for holobiont metrics or to do additional physiological measurements.
- Spin the S tube at 13,000 rpm on Labnet Spectrofuge 24D for 3 min.
- Transfer supertatant (1 mL) to a new tube labeled with teh coral ID + “H”. This is the host tube.
- Add 500 µL of PBS to the S tube and pipette up and down and vortex briefly to resuspend the symbiont pellet.
- Put both the H and S tubes in the freezer in respective “Host” and “Symbiont” boxes. Keep at -40°C.
- We have boxes “H1”, “H2”, “S1”, and “S2” to fit all our samples.
- Put the clean skeleton into a plastic cup and keep the whirlpack with the cup. Fill with 20-50% bleach to cover the skeleton. Leave in bleach for 8-10 hours.
- After bleaching, rinse the skeleton in fresh water and put in the drying oven at 80°C for 4-6 hours until dry.
- After dry, proceed with surface area (discussed below).
- Where any volumes or deviations in this protocol took place they were noted in metadata.
We followed this protocol for all samples over 2 days.
Airbrushing data can be found on GitHub here.
Note that two recruit samples (POC-R26 and ACR-R11) were very small and did not have any extra slurry in addition to the aliquot. For these samples, a subsample of 300 uL of resuspended symbiont pellet was removed and placed in a tube labeled with the coral ID and “Chl” to designate for future use for chlorophyll measurements. This is because the S tubes, which will be used for cell counting, will be thawed and therefore would compromise the chlorophyll quality.
First, we will use the S tube for cell density counting and complete surface area measurements. If Hollie or other members of the team have time, we can do host protein, chlorophyll content, or other analyses in the future.
Sunday - November 19, 2023
Airbrushing
We continued and finished airbrushing of all 90 samples today. See yesterday’s entry for protocol, notes, and data. Fragments from yesterday were placed in the drying oven at 08:00. Fragments from today were placed in bleach by 12:45 and put into the drying oven at 19:30.
Airbrushing data can be found on GitHub here.
Cell counting
We starting measuring symbiont cell densities on airbrushed samples S tubes today. Lucy and Ariana will both work on cell counting. We followed the E5 cell counting protocol. We did not make any modifications to this protocol.
Counts were done on a Neubauer Improved 0.1mm deep bright line hemocytometer (Hausser Scientific). We conducted 6 replicate counts per sample and calculated the CV (>90% of samples had CV below 10%). When CV was high, we repeated the counts. Data was entered daily in the notebook and is on GitHub here. Ariana and Lucy both worked on this until Ariana left on Nov 21. Lucy continued and finished the counts afterwards.
Surface area
We measured surface area of the corals airbrushed on Nov 18th. We followed the E5 surface area protocol. We are using the standard curve generated from surface area measurements on November 8, 2023. Samples weighed on a Mettler Toledo PB303-S/FACT scale.
Data were recorded in GitHub here.
Monday - November 20, 2023
Surface area
We measured surface area of the corals airbrushed on Nov 19th. Data were recorded in GitHub here. Dried skeletons were left in the molecular lab cabinet until data was analyzed.
Cleaning and inventory
Ariana cleaned the lab, organized supplies, and conducted sample inventory. Everything is done!
Cell counting
Lucy continued cell counting and will do a few samples each day until all are completed.
Tuesday - November 21, 2023
Today we attended the Fete de la Science festival in Papeete with Reva Atea!



